The work of Ketty Tamburrini, Sonia Longhi and their collaborators is highlighted by CNRS Biologie, congratulations!
A new mechanism involved in cellulose degradation by a phytopathogenic fungus
In an article published in the journal PNAS, scientists describe a mechanism used by a phytopathogenic fungus to “boost” cellulose degradation during plant infection. The discovery of this process could pave the way for the production of new enzymatic cocktails to degrade cellulose, a polymer abundant on Earth and a source of energy but highly resistant to degradation.
Plant pathogenic fungi (phytopathogens) have the ability to degrade plant cell walls containing cellulose during the infection process. Some of the enzymes involved in cellulose degradation have peculiarities that have caught the attention of scientists. Indeed, these enzymes have regions that are “intrinsically disordered”, meaning they lack a unique and precise three-dimensional structure. These regions, possessing a constantly changing structure, are nevertheless capable of exerting a biological function precisely because of this lack of fixed structure. Their malleability allows them to establish interactions with many partners, and this promiscuity enables them to regulate functions in numerous biological processes.
Studying a widespread fungal disease significantly impacting the yield of vegetable, fruit, and cereal crops
An exhaustive bioinformatic analysis of 27,060 sequences of these enzymes, called Lytic Polysaccharide MonoOxygenases (LPMOs), revealed that these disordered appendices were widespread among these enzymes. The prevalence of these disordered regions, whose function was unknown and neglected by the scientific community, in enzymes from highly divergent organisms in terms of evolution hinted at an important functional role. Scientists then undertook the study of these enigmatic regions in phytopathogenic fungi of the genus Colletotrichum, responsible for anthracnose, a widespread fungal disease significantly impacting the yield of vegetable, fruit, and cereal crops. To penetrate plant tissues, these fungi have evolved over hundreds of millions of years to acquire the ability to degrade polymers such as cellulose. They use specialized enzymes, such as LPMOs, for this purpose.
During this study, scientists focused on LPMOs belonging to the AA9 family of the fungus Colletotrichum orbiculare. Through a bioinformatics approach, regions predicted to be disordered at one end of these LPMOs were identified, and two of these enzymes were then selected for studying these enigmatic extensions. These two fungal enzymes were subsequently produced by recombinant means in yeast, an organism capable of manufacturing proteins similar to, or identical to, those produced by Colletotrichum. The disordered nature of these regions was confirmed using biophysical methods such as small-angle X-ray scattering, a technique particularly suitable for studying flexible systems.
New enzymatic cocktails to more effectively degrade cellulose
Unexpectedly, these enzymes were found to be capable of dimerization, and scientists demonstrated that this dimerization process confers enhanced properties to these enzymes in terms of substrate binding and cellulose activity. Dimerization relies on the formation of a disulfide bridge involving a cysteine residue located in the disordered extension. Thus, these disordered regions, which were previously systematically eliminated before any structural and functional study of LPMOs, turned out to be more than simple appendices and are essential for the formation of enzyme dimers with higher enzymatic activity compared to the monomeric form.
The results also show that the dimerization process is not a simple artifact occurring in a test tube containing purified enzyme, as this process also occurs in the fungus C. orbiculare, where this enzyme is indeed secreted in dimer form during infection. Scientists then deciphered the role of this enzyme in the plant penetration mechanism: eliminating this enzyme in C. orbiculare results in a defect in appressorium formation, the specialized structure for penetrating plant tissues.
Furthermore, scientists have shown that the ability to dimerize is not a unique property of LPMOs from C. orbiculare, and it is also observed in other fungi of the genus Colletotrichum. The findings from this study illustrate the ability of fungi to adapt to their environment by producing enzymes capable of degrading recalcitrant polymers and pave the way for the development of new enzymatic cocktails to more effectively degrade cellulose
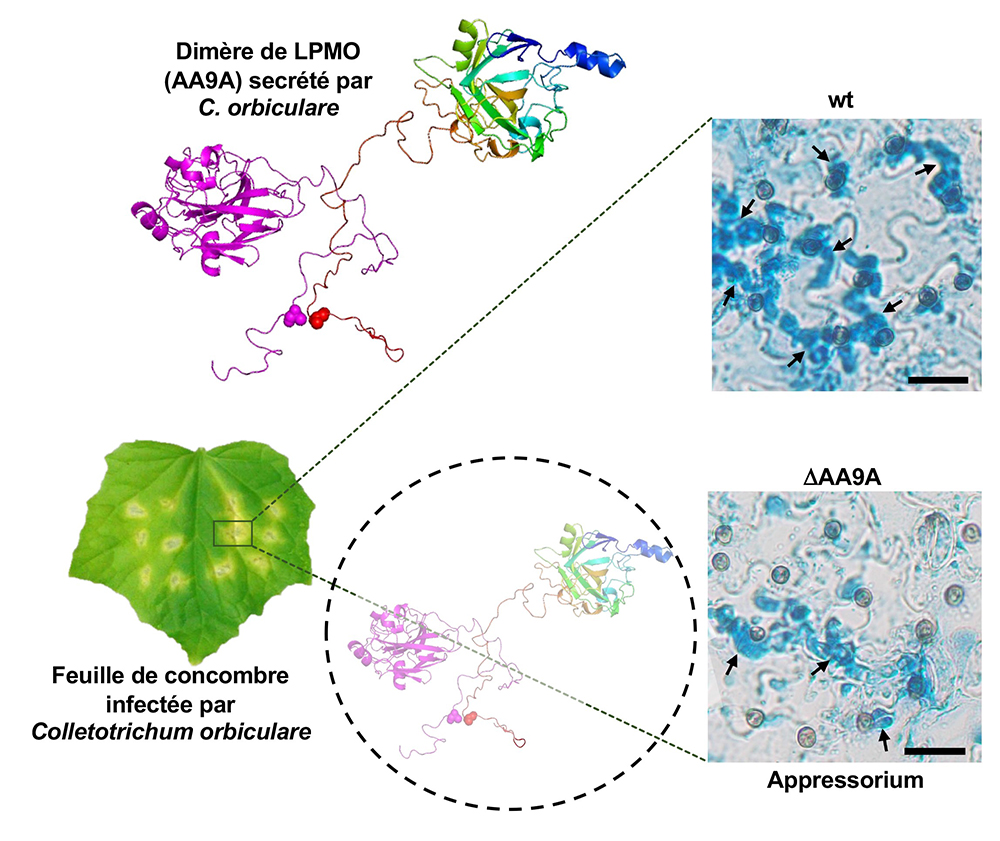
Figure : Feuille de concombre infectée par le champignon phytopathogène Colletotrichum orbiculare. Ce champignon secrète un cocktail d’enzymes parmi lesquelles figure l’enzyme AA9A, une LPMO de la famille AA9, capable de dégrader la cellulose, un polymère complexe présente à la surface des feuilles. Un conformère de cette enzyme à l’état dimérique est représenté en « cartoon » avec un monomère en couleur mauve (à gauche) et l’autre monomère en arc-en-ciel (à droite). Le résidu de cystéine, présent dans l’appendice C-terminal désordonné et impliqué dans la formation du pont disulfure entrainant la dimérisation de l’enzyme, est montré en sphères. L’infection de la plante par la souche sauvage de C. orbiculare (wt) possédant une LPMO AA9A intacte et fonctionnelle, permet une pénétration efficace de la plante, alors que chez la souche DAA9A, où l’enzyme LPMO AA9A a été éliminée, on observe un défaut dans la formation de l’appressorium et un nombre réduit de hyphes pénétratives (flèches noires).
En savoir plus :
The disordered C-terminal tail of fungal LPMOs from phytopathogens mediates protein dimerization and impacts plant penetration. Tamburrini KC, Kodama S, Grisel S, Haon M, Nishiuchi T, Bissaro B, Kubo Y, Longhi S, Berrin JG. Proc Natl Acad Sci U S A. 2024 Mar 26;121(13):e2319998121. doi: 10.1073/pnas.2319998121. Epub 2024 Mar 21. PMID: 38513096
Contact
Sonia Longhi
Directrice de recherche CNRS au laboratoire Architecture et fonction des macromolécules biologiques (AFMB)
Jean-Guy Berrin
Chercheur Inrae
Laboratoires
Architecture et fonction des macromolécules biologiques – AFMB (CNRS/Aix Marseille université)
163, Avenue de Luminy, Case 932
13288 Marseille
Biodiversité et Biotechnologie Fongiques – BBF (Inrae/Aix Marseille Université)
163, Avenue de Luminy, Case 925
13288 Marseille
Published on March 25, 2024



