Équipe
Réplicases Virales : Structure Mécanisme et Drug-Design
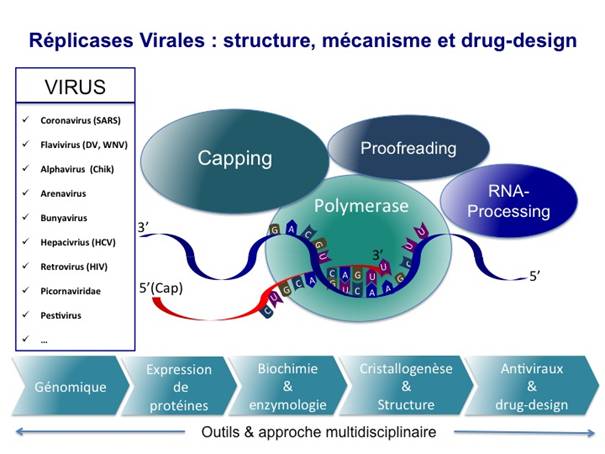
Notre équipe cherche à comprendre les mécanismes moléculaires de la réplication des virus émergents par la caractérisation des activités enzymatiques et l’analyse structurale des protéines formant le complexe de réplication/transcription. Ces études sont un préalable au développement d’inhibiteurs spécifiques de ces enzymes et devraient permettre le développement de nouvelles stratégies antivirales.
Les virus à génome ARN sont régulièrement associés à l’émergence de maladies infectieuses. Parmi les plus connues, les épidémies liées aux Bunyavirales (LCMV, Lassa, Vallée du Rift…), au Filovirus (Ebola), aux Nidovirales (Coronavirus), aux Flavivirus ( Chikungunya ou le virus de la Dengue) et aux maladies chroniques (HIV, HCV, HBV, HEV) illustrent des problèmes de santé publique générés à l’échelle mondiale.
L’équipe multidisciplinaire « Réplicases Virales : structure, mécanisme et drug–design » s’intéresse à l’ensemble des enzymes de virus émergents impliquées dans la réplication de leur génome et à la synthèse (transcription) de leurs ARN messagers. Ces enzymes sont le plus souvent associées au sein d’un complexe de réplication/transcription (RTC). Nos études portent non seulement sur les polymérases virales qui coordonnent la réplication virale, mais également sur les enzymes impliquées dans des mécanismes de modifications des ARNs (coiffage, édition, correction …) ou encore sur les protéines impliquées dans la régulation de la réplication.
Au delà de l’analyse restreinte au complexe réplicatif, nous nous nous intéressons également à l’interférence entre cette machinerie enzymatique avec les mécanismes de l’immunité innée de la cellule. Les enzymes de la réplication sont donc à double titre des cibles antivirales privilégiées et la compréhension de leur mode de fonctionnement participe à la conception de molécules à haut potentiel antiviral. Pour soutenir ces projets de recherche, le laboratoire s’appuie sur la plateforme de criblage d’inhibiteurs (PCML) et sur une banque de protéines virales recombinantes.
Etude des ARN polymérases ARN-dépendantes
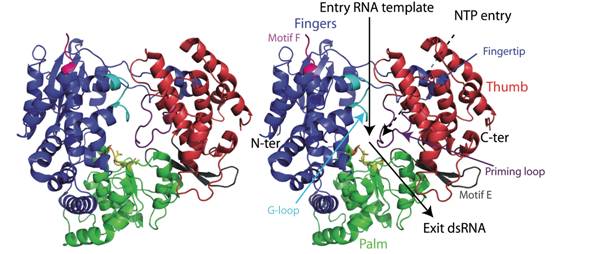
L’étude des polymérases constitue naturellement une des thématiques de recherche importante du laboratoire (HIV, HCV, dengue, West Nile, SARS, BVDV, Coxsackie virus). Nous disposons des outils permettant de suivre leur activité enzymatique. Nos travaux ont notamment permis ces dernières années de résoudre la structure de plusieurs enzymes, de déterminer des mécanismes de fonctionnement, et d’identifier les partenaires actifs dans les complexes multi-protéiques. Nous avons également développé des inhibiteurs originaux et étudié les mécanismes de résistance vis à vis de ces antiviraux.
Développement d’antiviraux
Dans la recherche de molécules antivirales, nous utilisons 2 stratégies de découverte : l’une basée sur la synthèse de molécules conçues rationnellement, grâce à la biochimie et la biologie structurale et l’autre basée sur l’optimisation de molécules « hits » issues de résultats de criblages. Dans ces deux approches, nos cibles sont des protéines du système réplicatif des virus. L’originalité et la qualité du travail qui est réalisé sur ces sujets tiennent en grande partie à la multidisciplinarité des projets combinant biochimie, biologie structurale, biologie moléculaire, bio-informatique, modélisation moléculaire, cristallographie, enzymologie, virologie et chimie.
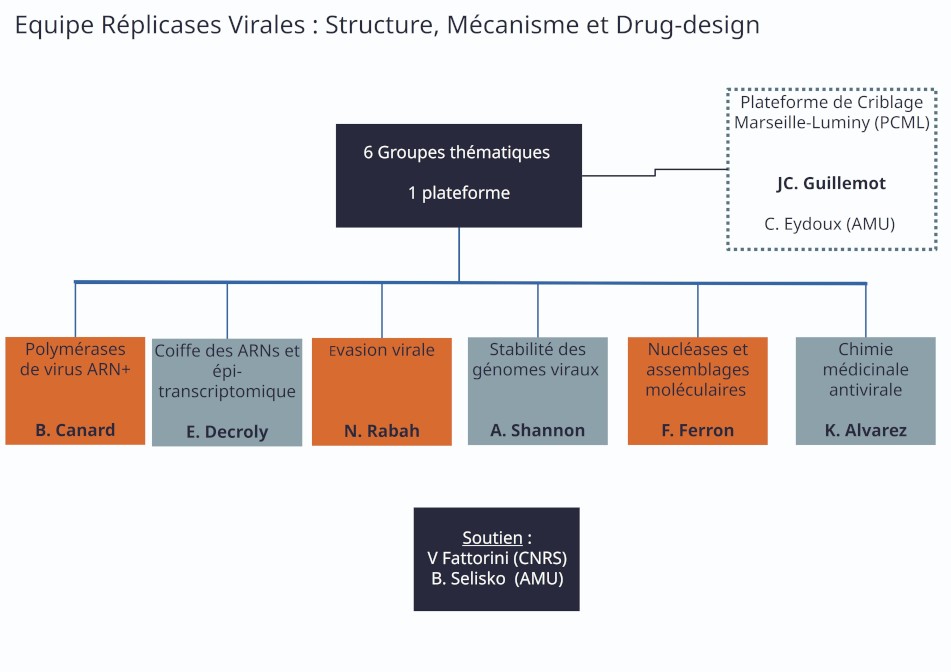
Organigramme de l’équipe
Domaines de recherche
- activités enzymatiques
- réplication/transcription
- stratégies antivirales
- ARN polymérases
- nucléases
- méthyltransférases
- protéases
- nucléoproteines
Membres
Publications
AIR-MT: AI-based design of RNA Methyltransferases for RNA-based therapeutics
Priscila Sutto, Bruno Canard, François Ferron
(2026)
Nuclear activities and interactome of the NS5 protein of Tick-Borne Encephalitis Virus
Maxime Chazal, Aïssatou Aïcha Sow, Elodie Le Seac’h, Dana Hawasheen, Margarida Bonifacio, Mikael Feracci, Ségolène Gracias, Adrià Sogues, Melissa Molho, Heng Pan, Boris Bonaventure, Jean-Pierre Quivy, Geneviève Almouzni, Etienne Decroly, Holly Ramage, Jeffrey Johnson, Vincent Caval, Nolwenn Jouvenet
Can plitidepsin be used as an antiviral against RSV?
Charlotte Estampes, Jenna Fix, Julien Sourimant, Priscila Sutto-Ortiz, Charles-Adrien Richard, Etienne Decroly, Marie Galloux, Jean-François Eléouët
MSphere 10 (2025)10.1128/msphere.00127-25
Publications de l'équipe
AIR-MT: AI-based design of RNA Methyltransferases for RNA-based therapeutics
Priscila Sutto, Bruno Canard, François Ferron
(2026)
Nuclear activities and interactome of the NS5 protein of Tick-Borne Encephalitis Virus
Maxime Chazal, Aïssatou Aïcha Sow, Elodie Le Seac’h, Dana Hawasheen, Margarida Bonifacio, Mikael Feracci, Ségolène Gracias, Adrià Sogues, Melissa Molho, Heng Pan, Boris Bonaventure, Jean-Pierre Quivy, Geneviève Almouzni, Etienne Decroly, Holly Ramage, Jeffrey Johnson, Vincent Caval, Nolwenn Jouvenet
Can plitidepsin be used as an antiviral against RSV?
Charlotte Estampes, Jenna Fix, Julien Sourimant, Priscila Sutto-Ortiz, Charles-Adrien Richard, Etienne Decroly, Marie Galloux, Jean-François Eléouët
MSphere 10 (2025)10.1128/msphere.00127-25
Crystallographic characterisation and development of bi-substrate inhibitors of coronavirus nsp14 methyltransferase
Irene Georgiou, Colin Robinson, Sean O’byrne, Alex Matsuda, Przemysław Grygier, Craig D Smith, Sandra O’neill, Shamshad A Ahmad, Suzanne Norval, John M Post, G J Mirjam Groenewold, Nadya Urakova, Patrick Wanningen, Leanid Kresik, Jacek Plewka, Adrien Delpal, Kexin See, Thomas Eadsforth, Kinga Wierzbicka, Etienne Decroly, Kumar Singh Saikatendu, Edcon Chang, Eric J Snijder, Krzysztof Pyrć, Anna Czarna, Duncan Scott, Ian H Gilbert
Synthesis and in vitro inhibitory activity of N-arylsulfonamide adenosine analogues designed to target SARS-CoV-2 nsp14 N7-methyltransferase
Marcel Hausdorff, Adrien Delpal, Hugo Machin, Amina Tahir, Jim Zoladek, Floriane Gucciardi, Jitendriya Swain, Nathalie Gros, Delphine Muriaux, Sébastien Nisole, Bruno Canard, Jean-Jacques Vasseur, Etienne Decroly, Françoise Debart
Bioorganic Chemistry 166:109104 (2025)10.1016/j.bioorg.2025.109104
Direct interaction between RSV polymerase L and active Rab11a mediates viral ribonucleoprotein transport to assembly sites
Claire Giry, Ana Joaquina Jimenez, Julie de Oliveira, Fatemeh Okhravijouybari, Mélissa Bessonne, Anne Huard De Verneuil, Patrick England, Priscila Sutto-Ortiz, Didier Chevret, Amina Tahir, Françoise Debart, Etienne Decroly, Julien Sourimant, Marie-Anne Rameix-Welti
Expériences en virologie. Bénéfices et risques par Etienne Decroly
Dominique Desbois
Le Monde Diplomatique 25 p. (2025)
Mapping the impact of 1'-, 2'-and 4'-nucleotide modifications on the Respiratory Syncytial Virus RNA-dependent RNA polymerase
Priscila Sutto-Ortiz, Barbara Selisko, François Ferron, Jean-Pierre Sommadossi, Adel Moussa, Steven Good, Bruno Canard, Etienne Decroly
Antiviral Research (2025)10.1016/j.antiviral.2025.106298
RABV L protein plays a role in immune escape through its methyltransferase activity
Marilys Castet, Wahiba Aouadi, Lauriane Kergoat, Ambre Bulteau, Rachel Legendre, Rania Ouazahrou, Lena Feige, Hugo Varet, Aurore Attina, Christophe Hirtz, Alexandre David, Etienne Decroly, Hervé Bourhy, Florence Larrous
Structural Insights into the Protein Mannosyltransferase from Mycobacterium tuberculosis reveal a WW-Domain-Like Protein Motif in Bacteria
Nicolas Géraud, Chloé Rivière, Camille Falcou, Gianluca Cioci, Carine Froment, Virginie Gervais, Julien Marcoux, Martine Gilleron, Jérôme Nigou, Emeline Fabre, Michel Rivière
Communications Biology 8:1175 (2025)10.1038/s42003-025-08593-9
Can Plitidepsin Be Used as an Antiviral Against RSV?
Charlotte Estampes, Jenna Fix, Julien Sourimant, Priscila Sutto-Ortiz, Charles-Adrien Richard, Etienne Decroly, Marie Galloux, Jean-François Eléouët
Rapport 25-06. De l’origine du SARS-CoV-2 aux risques de zoonoses et de manipulations dangereuses de virus
Christine Rouzioux, Etienne Decroly, Patrick Berche, Patrice Binder, François Bricaire, Yves Buisson, Bernard Charpentier
650-662 (2025)10.1016/j.banm.2025.04.016
Neurotoxic Implications of Human Coronaviruses in Neurodegenerative Diseases: A Perspective from Amyloid Aggregation
Thi Hong Van Nguyen, Francois Ferron, Kazuma Murakami
ACS Chemical Biology 20:983-992 (2025)10.1021/acschembio.5c00153
Analysis of substrate recognition by Mycobacterium protein-mannosyl transferase reveals WW-domain-like recognition in bacteria
Michel Rivière, Nicolas Géraud, Chloé Rivière, Camille Falcou, Martine Gilleron, Carine Froment, Julien Marcoux, Gianluca Cioci, Jérôme Nigou, Emeline Fabre
Derivatives of MOPS : promising scaffolds for SARS coronaviruses Macro domain‐targeted inhibition
Oney Ortega Granda, Karine Alvarez, Benjamin Morin, Bruno Canard, François Ferron, Nadia Rabah
FEBS Journal 292:2865-2881 (2025)10.1111/febs.70039
A coronavirus assembly inhibitor that targets the viral membrane protein
Manon Laporte, Dirk Jochmans, Dorothée Bardiot, Lowiese Desmarets, Oliver Debski-Antoniak, Giulia Mizzon, Rana Abdelnabi, Pieter Leyssen, Winston Chiu, Zhikuan Zhang, Norimichi Nomura, Sandro Boland, Umeharu Ohto, Yannick Stahl, Jurgen Wuyts, Steven de Jonghe, Annelies Stevaert, Martijn van Hemert, Brenda Bontes, Patrick Wanningen, G. Groenewold, Aneta Zegar, Katarzyna Owczarek, Sanjata Joshi, Mohamed Koukni, Philippe Arzel, Hugo Klaassen, Jean-Christophe Vanherck, Ilse Vandecaetsbeek, Niels Cremers, Kim Donckers, Thibault Francken, Tina van Buyten, Jasper Rymenants, Joost Schepers, Krzysztof Pyrc, Rolf Hilgenfeld, Jean Dubuisson, Berend-Jan Bosch, Frank van Kuppeveld, Cecilia Eydoux, Etienne Decroly, Bruno Canard, Lieve Naesens, Birgit Weynand, Eric Snijder, Sandrine Belouzard, Toshiyuki Shimizu, Ralf Bartenschlager, Daniel Hurdiss, Arnaud Marchand, Patrick Chaltin, Johan Neyts
Nature 640:514-523 (2025)10.1038/s41586-025-08773-x
Nucleotide analogues and mpox: Repurposing the repurposable
Ashleigh Shannon, Bruno Canard
Antiviral Research 234:106057 (2025)10.1016/j.antiviral.2024.106057
Design, synthesis and evaluation of arylpurine-based sinefungin mimetics as zika virus methyltransferase inhibitors
Natalia del Río, Iván Arribas-Álvarez, José-María Orduña, Priscila Sutto-Ortiz, Johan Neyts, Suzanne Kaptein, Etienne Decroly, Eva-María Priego, María-Jesús Pérez-Pérez
RSC Advances 15:37309-37324 (2025)10.1039/d5ra05362e
Comment le virus Usutu résiste à la restriction par l’exonucléase ISG20
Jim Zoladek, Vincent Caval, Jean-Christophe Paillart, Etienne Decroly, Sébastien Nisole
Médecine/Sciences 40:973-975 (2024)10.1051/medsci/2024166
The effects of Remdesivir's functional groups on its antiviral potency and resistance against the SARS-CoV-2 polymerase
Bhawna Sama, Barbara Selisko, Camille Falcou, Véronique Fattorini, Géraldine Piorkowski, Franck Touret, Kim Donckers, Johan Neyts, Dirk Jochmans, Ashleigh Shannon, Bruno Coutard, Bruno Canard
Antiviral Research 232 (2024)10.1016/j.antiviral.2024.106034
A specific domain within the 3′ untranslated region of Usutu virus confers resistance to the exonuclease ISG20
Jim Zoladek, Priscila El Kazzi, Vincent Caval, Valérie Vivet-Boudou, Marion Cannac, Emma Davies, Soléna Rossi, Inès Bribes, Lucile Rouilly, Yannick Simonin, Nolwenn Jouvenet, Etienne Decroly, Jean-Christophe Paillart, Sam J Wilson, Sébastien Nisole
Nature Communications 15:8528 (2024)10.1038/s41467-024-52870-w
Alphavirus nsP3 organizes into tubular scaffolds essential for infection and the cytoplasmic granule architecture
Vasiliya Kril, Michael Hons, Celine Amadori, Claire Zimberger, Laurine Couture, Yara Bouery, Julien Burlaud-Gaillard, Andrei Karpov, Denis Ptchelkine, Alexandra Thienel, Beate Kümmerer, Ambroise Desfosses, Rhian Jones, Philippe Roingeard, Laurent Meertens, Ali Amara, Juan Reguera
Nature Communications 15:8106 (2024)10.1038/s41467-024-51952-z
When 2′-O-methylation throws a wrench in HIV-1 reverse transcriptase
Alice Decombe, Olve Peersen, Etienne Decroly
Virologie 28:277-293 (2024)10.1684/vir.2024.1056
The Phlebovirus Ribonucleoprotein: An Overview
François Ferron, Julien Lescar
2824:259-280 (2024)10.1007/978-1-0716-3926-9_17
JNJ-7184, a respiratory syncytial virus inhibitor targeting the connector domain of the viral polymerase
Brecht Bonneux, Afzaal Shareef, Sergey Tcherniuk, Brandon Anson, Suzanne de Bruyn, Nick Verheyen, Kim Thys, Nádia Conceição-Neto, Marcia van Ginderen, Leen Kwanten, Nina Ysebaert, Luc Vranckx, Elien Peeters, Ellen Lanckacker, Jack M Gallup, Panchan Sitthicharoenchai, Sarhad Alnajjar, Mark R Ackermann, Suraj Adhikary, Anusarka Bhaumik, Aaron Patrick, Amy Fung, Priscila Sutto-Ortiz, Etienne Decroly, Stephen W Mason, David Lançois, Jerome Deval, Zhinan Jin, Jean-François Éléouët, Rachel Fearns, Anil Koul, Dirk Roymans, Peter Rigaux, Florence Herschke
Antiviral Research 227:105907 (2024)10.1016/j.antiviral.2024.105907
Il est urgent d’harmoniser les réglementations internationales de biosécurité dans la recherche
Etienne Decroly
Le monde (2024)
How did you annotate your new viral genome without it ? VAZyMolO-2
Vincent Wilde, Bruno Canard, Francois Ferron
(2024)
Identification of adenosine analogues that inhibit the N7 methyltransferase activity of SARS-CoV-2
Adrien Delpal, Marcel Hausdorff, Sarah Barelier, Jean-Jacques Vasseur, Etienne Decroly, Françoise Debart
(2024)
Effets de la 2′-O-méthylation de l’ARN génomique du VIH-1 sur la réplication virale
Alice Decombe, Priscila El-Kazzi, Sébastien Nisole, Étienne Decroly
Médecine/Sciences 40:421-427 (2024)10.1051/medsci/2024046
Biophysical and structural study of La Crosse virus endonuclease inhibition for the development of new antiviral options
Mikael Feracci, Sergio Hernandez, Laura Garlatti, Clemence Mondielli, Renaud Vincentelli, Bruno Canard, Juan Reguera, François Ferron, Karine Alvarez
International Union of Crystallography journal 11:374-383 (2024)10.1107/S205225252400304X
La mutation P323L de nsp12 de SARS-CoV-2est-elle un marqueur d’espèce hôte?
Camille Falcou, Cécilia Eydoux, Léa Lo-Belo, Eric Snijder, Bruno Canard
(2024)
Utilisation de conjugués ARN/SAM en tant qu'inhibiteurs de 2'O méthyltransférases virales
Adrien Delpal, Rostom Ahmed-Belkacem, Priscila Sutto-Ortiz, Joris Troussier, Bruno Canard, Jean- Jacques Vasseur, Étienne Decroly, Françoise Debart
(2024)
Modifications épitranscriptomiques des ARN viraux : effets pro-et anti-viraux des 2'-Ométhylations des génomes viraux
Etienne Decroly, Alice Decombe, Priscila El Kazzi, Sébastien Nisole
(2024)
The Journées francophones de virologie go international with a visit to Brussels
Anne Op de Beeck, Etienne Decroly
28:59-60 (2024)10.1684/vir.2024.1042
The activation chain of the broad-spectrum antiviral bemnifosbuvir at atomic resolution
Aurelie Chazot, Claire Zimberger, Mikael Feracci, Adel Moussa, Steven Good, Jean-Pierre Sommadossi, Karine Alvarez, Francois Ferron, Bruno Canard
PLoS Biology (2024)10.1371/journal.pbio.3002743
5′-cap RNA/SAM mimetic conjugates as bisubstrate inhibitors of viral RNA cap 2′-O-methyltransferases
Rostom Ahmed‐belkacem, Priscila Sutto-Ortiz, Adrien Delpal, Joris Troussier, Bruno Canard, Jean-Jacques Vasseur, Etienne Decroly, Françoise Debart
Bioorganic Chemistry 143:107035 (2024)10.1016/j.bioorg.2023.107035
N-arylsulfonamide-based adenosine analogues to target RNA cap N7-methyltransferase nsp14 of SARS-CoV-2
Rostom Ahmed‐belkacem, Joris Troussier, Adrien Delpal, Bruno Canard, Jean-Jacques Vasseur, Etienne Decroly, Françoise Debart
RSC Medicinal Chemistry (2024)10.1039/d3md00737e
Structural flexibility of Toscana virus nucleoprotein in the presence of a single-chain camelid antibody
Nicolas Papageorgiou, Amal Baklouti, Julie Lichière, Aline Desmyter, Bruno Canard, Bruno Coutard, François Ferron
Acta crystallographica Section D : Structural biology [1993-...] 80 (2024)10.1107/S2059798324000196
Synthesis of acyclic analogues of adenosine sulfonamides and their activity against RNA cap guanine N 7-methyltransferase of SARS-CoV-2
Rostom Ahmed‐belkacem, Adrien Delpal, Bruno Canard, Jean-Jacques Vasseur, Etienne Decroly, Françoise Debart
New Journal of Chemistry 143:107035 (2024)10.1039/D4NJ02481H
Identification of Novel Non-Nucleoside Inhibitors of Zika Virus NS5 Protein Targeting MTase Activity
Diego Fiorucci, Micaela Meaccini, Giulio Poli, Maria Alfreda Stincarelli, Chiara Vagaggini, Simone Giannecchini, Priscila Sutto-Ortiz, Bruno Canard, Etienne Decroly, Elena Dreassi, Annalaura Brai, Maurizio Botta
International Journal of Molecular Sciences 25:2437 (2024)10.3390/ijms25042437
Single-cell RNA-sequencing of PBMCs from SAVI patients reveals disease-associated monocytes with elevated integrated stress response
Camille de Cevins, Laure Delage, Maxime Batignes, Quentin Riller, Marine Luka, Anne Remaury, Boris Sorin, Tinhinane Fali, Cécile Masson, Bénédicte Hoareau, Catherine Meunier, Mélanie Parisot, Mohammed Zarhrate, Brieuc P Pérot, Víctor García-Paredes, Francesco Carbone, Lou Galliot, Béatrice Nal, Philippe Pierre, Luc Canard, Charlotte Boussard, Etienne Crickx, Jean-Claude Guillemot, Brigitte Bader-Meunier, Alexandre Bélot, Pierre Quartier, Marie-Louise Frémond, Bénédicte Neven, Galina Boldina, Franck Augé, Fischer Alain, Michel Didier, Frédéric Rieux-Laucat, Mickaël M Ménager
Cell Reports Medicine 4:101333 (2023)10.1016/j.xcrm.2023.101333
Structural and mechanistic insights into the inhibition of respiratory syncytial virus polymerase by a non-nucleoside inhibitor
Xiaodi Yu, Pravien Abeywickrema, Brecht Bonneux, Ishani Behera, Brandon Anson, Edgar Jacoby, Amy Fung, Suraj Adhikary, Anusarka Bhaumik, Rodrigo Carbajo, Suzanne de Bruyn, Robyn Miller, Aaron Patrick, Quyen Pham, Madison Piassek, Nick Verheyen, Afzaal Shareef, Priscila Sutto-Ortiz, Nina Ysebaert, Herman van Vlijmen, Tim Jonckers, Florence Herschke, Jason Mclellan, Etienne Decroly, Rachel Fearns, Sandrine Grosse, Dirk Roymans, Sujata Sharma, Peter Rigaux, Zhinan Jin
Communications Biology 6:1074 (2023)10.1038/s42003-023-05451-4
Internal RNA 2′-O-methylations on HIV-1 genome impair reverse transcription.
Alice Decombe, Olve Peersen, Priscila Sutto-Ortiz, Célia Chamontin, Géraldine Piorkowski, Bruno Canard, Sébastien Nisole, Etienne Decroly
Nucleic Acids Research (2023)
Internal RNA 2′- O -methylation on the HIV-1 genome impairs reverse transcription
Alice Decombe, Olve Peersen, Priscila Sutto-Ortiz, Célia Chamontin, Géraldine Piorkowski, Bruno Canard, Sébastien Nisole, Etienne Decroly
Nucleic Acids Research 52:1359-1373 (2023)10.1093/nar/gkad1134
BIOCHEMICAL AND STRUCTURAL CHARACTERIZATION OF L POLYMERASE PROTEIN FROM LCMV
Candice Sartre, Sergio Hernández, Barbara Selisko, Denis Ptchelkine, Bruno Canard, Karine Alvarez, Francois Ferron
(2023)
Tick‐borne flavivirus NS5 antagonizes interferon signaling by inhibiting the catalytic activity of TYK2
Ségolène Gracias, Maxime Chazal, Alice Decombe, Yves Unterfinger, Adrià Sogues, Lauryne Pruvost, Valentine Robert, Sandrine Lacour, Manon Lemasson, Marion Sourisseau, Zhi Li, Jennifer Richardson, Sandra Pellegrini, Etienne Decroly, Vincent Caval, Nolwenn Jouvenet
EMBO Reports 24:e57424 (2023)10.15252/embr.202357424
Development of a novel target-based cell assay, reporter of the activity of Mycobacterium tuberculosis protein- O -mannosyltransferase
Nicolas Géraud, Camille Falcou, Julien Parra, Carine Froment, David Rengel, Odile Burlet-Schiltz, Julien Marcoux, Jérôme Nigou, Michel Rivière, Emeline Fabre
Glycobiology 33:1139-1154 (2023)10.1093/glycob/cwad072
Structural characterisation and inhibition of Arenavirus replication complex elements : assembly, function and inhibition of embedded nucleases
Sergio Hernández, Nicolas Papageorgiou, Maria Spiliopoulou, Mikael Feracci, Laura Garlatti, Barbara Selisko, Afroditi Vaitsopoulou, Thi-Hong van Nguyen, Clemence Mondielli, Magali Saez-Ayala, Elsie Yekwa Laban, Irene Margiolaki, B. Canard, Karine Alvarez, Francois Ferron
(2023)
Structural characterisation and inhibition of arenavirus replication complex elements: assembly, function and inhibition of embedded nucleases
S. Hernandez, N. Papageorgiou, M. Spiliopoulou, M. Ferracci, L. Garlatti, B. Selisko, T.-H. Nguyen, C. Mondielli, M. Saez-Ayala, Y. Laban, I. Margiolaki, B. Canard, Karine Alvarez, Francois Ferron
79:C677-C677 (2023)10.1107/S2053273323089404
Cellular activation pathway of bemnifosbuvir (AT-527), a drug candidate against SARS-CoV-2 infections
A. Chazot, C. Zimberger, M. Feracci, S. Hernandez, C. Falcou, A. Moussa, S. Good, J.-P. Sommadossi, Francois Ferron, Karine Alvarez, B. Canard
79:C1042-C1042 (2023)10.1107/S2053273323085790
Viral Instant Mutation Viewer (VIMVer) : a tool to speed up the identification and analysis of new SARS-CoV2 emerging variant and beyond
Vincent Wilde, Bruno Canard, François Ferron
Viruses 15:1628 (2023)10.3390/v15081628
Structural and functional characterization of the Protein-O-MannosylTransferase of Mycobacterium tuberculosis MtPMT : a potential target of innovative therapeutic inhibitors
Chloé Rivière, Nicolas Géraud, Camille Falcou, Emeline Fabre, Michel Rivière
(2023)
Protéine 2C d’Entérovirus : protéine clé de la réplication virale et cible antivirale
Priscila El Kazzi, Carole Yaacoub, Ziad Fajloun, Patrice Vanelle, Etienne Decroly, Bruno Coutard, Karine Barral
Virologie 27:173-188 (2023)10.1684/vir.2023.1001
Le complexe nsp14:nsp10 du SARS-CoV-2 stabilisé par un nanobody
Pierre Gauffre, Anaïs Gaubert, Maria Mate-Perez, Bruno Canard, François Ferron
27:48-49 (2023)10.1684/vir.2023.993
Identification d'analogues d'adénosine, inhibiteurs de l'activité méthyltransférase du SARS-CoV-2
Adrien Delpal, Marcel Hausdorff, Sarah Barelier, Jean-Jacques Vasseur, Bruno Canard, Étienne Decroly, Françoise Debart
(2023)
Protein-primed RNA synthesis in SARS-CoVs and structural basis for inhibition by AT-527
Ashleigh Shannon, Véronique Fattorini, Bhawna Sama, Barbara Selisko, Mikael Feracci, Camille Falcou, Pierre Gauffre, Priscila El Kazzi, Etienne Decroly, Nadia Rabah, Karine Alvarez, Cécilia Eydoux, Jean-Claude Guillemot, Françoise Debart, Jean-Jacques Vasseur, Mathieu Noel, Adel Moussa, Steven Good, Kai Lin, Jean-Pierre Sommadossi, Yingxiao Zhu, Xiaodong Yan, Hui Shi, François Ferron, Bruno Canard
Structural basis and dynamics of Chikungunya alphavirus RNA capping by nsP1 capping pores
Rhian Jones, Michael Hons, Nadia Rabah, Noelia Zamarreño, Rocío Arranz, Juan Reguera
Proceedings of the National Academy of Sciences of the United States of America 120 (2023)10.1073/pnas.2213934120
Discovery of a PDZ Domain Inhibitor Targeting the Syndecan/Syntenin Protein–Protein Interaction: A Semi-Automated “Hit Identification-to-Optimization” Approach
Laurent Hoffer, Manon Garcia, Raphael Leblanc, Mikael Feracci, Stéphane Betzi, Khaoula Ben Yaala, Avais Daulat, Pascale Zimmermann, Philippe Roche, Karine Barral, Xavier Morelli
Journal of Medicinal Chemistry (2023)10.1021/acs.jmedchem.2c01569
Interplay of RNA 2′-O-methylations with viral replication
Alice Decombe, Priscila El Kazzi, Etienne Decroly
Current Opinion in Virology 59:101302 (2023)10.1016/j.coviro.2023.101302
Design and synthesis of naturally-inspired SARS-CoV-2 inhibitors
Haitham Hassan, Jeanne Chiavaralli, Afnan Hassan, Loay Bedda, Tim Krischuns, Kuang-Yu Chen, Alice Shi Ming Li, Adrien Delpal, Etienne Decroly, Masoud Vedadi, Nadia Naffakh, Fabrice Agou, Sergio Mallart, Reem Arafa, Paola Arimondo
RSC Medicinal Chemistry 14:507-519 (2023)10.1039/D2MD00149G
Biochemistry of the Respiratory Syncytial Virus L Protein Embedding RNA Polymerase and Capping Activities
Priscila Sutto-Ortiz, Jean-François Eléouët, François Ferron, Etienne Decroly
Viruses 15:341 (2023)10.3390/v15020341
Notch activation shifts the fate decision of senescent progenitors toward myofibrogenesis in human adipose tissue
Nathalie Boulet, Anaïs Briot, Valentin Jargaud, David Estève, Anne Rémaury, Chloé Belles, Pénélope Viana, Jessica Fontaine, Lucie Murphy, Catherine Déon, Marie Guillemot, Catherine Pech, Yaligara Veeranagouda, Michel Didier, Pauline Decaunes, Etienne Mouisel, Christian Carpéné, Jason S Iacovoni, Alexia Zakaroff-Girard, Jean-Louis Grolleau, Jean Galitzky, Séverine Ledoux, Jean-Claude Guillemot, Anne Bouloumié
Aging Cell e13776 (2023)10.1111/acel.13776
Macro1 domain residue F156: A hallmark of SARS-CoV-2 de-MARylation specificity
Oney Ortega Granda, Karine Alvarez, Maria J Mate-Perez, Bruno Canard, François Ferron, Nadia Rabah
Virology 587:109845 (2023)10.1016/j.virol.2023.109845
AT-752 targets multiple sites and activities on the Dengue virus replication enzyme NS5
Mikael Feracci, Cécilia Eydoux, Véronique Fattorini, Lea Lo Bello, Pierre Gauffre, Barbara Selisko, Priscila Sutto-Ortiz, Ashleigh Shannon, Hongjie Xia, Pei-Yong Shi, Mathieu Noel, Françoise Debart, Jean-Jacques Vasseur, Steve Good, Kai Lin, Adel Moussa, Jean-Pierre Sommadossi, Aurélie Chazot, Karine Alvarez, Jean-Claude Guillemot, Etienne Decroly, François Ferron, Bruno Canard
Antiviral Research 212:105574 (2023)10.1016/j.antiviral.2023.105574
Amino Acid Polymorphisms on the Brazilian Strain of Yellow Fever Virus Methyltransferase Are Related to the Host’s Immune Evasion Mediated by Type I Interferon
Nathália Dias Furtado, Iasmim Silva de Mello, Andre Schutzer de Godoy, Gabriela Dias Noske, Glaucius Oliva, Bruno Canard, Etienne Decroly, Myrna Bonaldo
Viruses 15:191 (2023)10.3390/v15010191
Structure-guided optimization of adenosine mimetics as selective and potent inhibitors of coronavirus nsp14 N7-methyltransferases
Marcel Hausdorff, Adrien Delpal, Sarah Barelier, Laura Nicollet, Bruno Canard, Franck Touret, Agathe Colmant, Bruno Coutard, Jean-Jacques Vasseur, Etienne Decroly, Françoise Debart
European Journal of Medicinal Chemistry (2023)10.1016/j.ejmech.2023.115474
2C protein of Enterovirus: key protein of viral replication and antiviral target
Priscila El Kazzi, Carole Yaacoub, Ziad Fajloun, Patrice Vanelle, Etienne Decroly, Bruno Coutard, Karine Barral
Virologie 27:35-49 (2023)10.1684/vir.2023.1002
Internal RNA 2′O-methylation in the HIV-1 genome counteracts ISG20 nuclease-mediated antiviral effect
Priscila El Kazzi, Nadia Rabah, Célia Chamontin, Lina Poulain, François Ferron, Françoise Debart, Bruno Canard, Dorothée Missé, Bruno Coutard, Sébastien Nisole, Etienne Decroly
Nucleic Acids Research (2022)10.1093/nar/gkac996
A second type of N7-guanine RNA cap methyltransferase in an unusual locus of a large RNA virus genome
Ashleigh Shannon, Bhawna Sama, Pierre Gauffre, Théo Guez, Françoise Debart, Jean-Jacques Vasseur, Etienne Decroly, Bruno Canard, François Ferron
Nucleic Acids Research 50:11186-11198 (2022)10.1093/nar/gkac876
The SARS-CoV nsp12 Polymerase Active Site Is Tuned for Large-Genome Replication
Grace Campagnola, Vishnu Govindarajan, Annelise Pelletier, Bruno Canard, Olve Peersen
Journal of Virology 96 (2022)10.1128/jvi.00671-22
A highly specific N7-guanine RNA cap methyltransferase in an unusual locus of large RNA virus genome
A. Shannon, B. Sama, P. Gauffre, B. Makem-Tamekem, E. Decroly, B. Canard, Francois Ferron
Acta Crystallographica Section A : Foundations and Advances [2014-...] 78:e324-e324 (2022)10.1107/S2053273322094049
Identification of potent inhibitors of arenavirus and SARS-CoV-2 exoribonucleases by fluorescence polarization assay
Sergio Hernández, Mikael Feracci, Carolina Trajano de Jesus, Priscila El Kazzi, Rafik Kaci, Laura Garlatti, Clemence Mondielli, Fabrice Bailly, Philippe Cotelle, Franck Touret, Xavier de Lamballerie, Bruno Coutard, Etienne Decroly, Bruno Canard, François Ferron, Karine Alvarez
Antiviral Research 204:105364 (2022)10.1016/j.antiviral.2022.105364
A Versatile Class of 1,4,4-Trisubstituted Piperidines Block Coronavirus Replication In Vitro
Sonia de Castro, Annelies Stevaert, Miguel Maldonado, Adrien Delpal, Julie Vandeput, Benjamin van Loy, Cecilia Eydoux, Jean-Claude Guillemot, Etienne Decroly, Federico Gago, Bruno Canard, Maria-Jose Camarasa, Sonsoles Velázquez, Lieve Naesens
Pharmaceuticals 15:1021 (2022)10.3390/ph15081021
Arenaviridae NP-exonuclease inhibition by bisphosphonate
Thi Hong van Nguyen, Elsie Yekwa, Barbara Selisko, Bruno Canard, Karine Alvarez, Francois Ferron
International Union of Crystallography journal 9 (2022)10.1107/S2052252522005061
Structural and functional characterization of the Protein-O-MannosylTransferase of Mycobacterium tuberculosis
Chloé Rivière, Nicolas Géraud, Camille Falcou, Émeline Fabre, Michel Rivière
(2022)
A highly sensitive cell-based luciferase assay for high-throughput automated screening of SARS-CoV-2 nsp5/3CLpro inhibitors
K.Y. Chen, T. Krischuns, L. Ortega Varga, E. Harigua-Souiai, S. Paisant, A. Zettor, J. Chiaravalli, A. Delpal, D. Courtney, A. O'Brien, S.C. Baker, Etienne Decroly, C. Isel, F. Agou, Y. Jacob, A. Blondel, Nadia Naffakh
Antiviral Research 201:105272 (2022)10.1016/j.antiviral.2022.105272
Potent Inhibition of SARS-CoV-2 nsp14 N 7-Methyltransferase by Sulfonamide-Based Bisubstrate Analogues
Rostom Ahmed‐belkacem, Marcel Hausdorff, Adrien Delpal, Priscila Sutto-Ortiz, Agathe M G Colmant, Franck Touret, Natacha S Ogando, Eric J Snijder, Bruno Canard, Bruno Coutard, Jean-Jacques Vasseur, Etienne Decroly, Françoise Debart
Journal of Medicinal Chemistry 65:6231-6249 (2022)10.1021/acs.jmedchem.2c00120
Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity
Rachid Essalmani, Jaspreet Jain, Delia Susan-Resiga, Ursula Andréo, Alexandra Evagelidis, Rabeb Mouna Derbali, David Huynh, Frédéric Dallaire, Mélanie Laporte, Adrien Delpal, Priscila Sutto-Ortiz, Bruno Coutard, Claudine Mapa, Keith Wilcoxen, Etienne Decroly, Tram Nq Pham, Éric Cohen, Nabil Seidah
Journal of Virology 96 (2022)10.1128/jvi.00128-22
Mise au point d'une méthode de screening d'inhibiteurs de la Main protéase (nsp5) du Sars-CoV-2
Adrien Delpal, Valentine Robert, Bruno Coutard, Étienne Decroly, Nadia Naffakh
(2022)
Viral Instant Mutation Viewer (VIMVer): a tool to speed up the identification and analysis of new SARS-CoV2 emerging variant
Vincent Wilde, Bruno Canard, Francois Ferron
(2022)
A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase
Ashleigh Shannon, Véronique Fattorini, Bhawna Sama, Barbara Selisko, Mikael Feracci, Camille Falcou, Pierre Gauffre, Priscila El Kazzi, Adrien Delpal, Etienne Decroly, Karine Alvarez, Cécilia Eydoux, Jean-Claude Guillemot, Adel Moussa, Steven S Good, Paolo La Colla, Kai Lin, Jean-Pierre Sommadossi, Yingxiao Zhu, Xiaodong Yan, Hui Shi, Francois Ferron, Bruno Canard
Nature Communications 13 (2022)10.1038/s41467-022-28113-1
Fluoxetine targets an allosteric site in the enterovirus 2C AAA+ ATPase and stabilizes a ring-shaped hexameric complex
Daniel L Hurdiss, Priscila El Kazzi, Lisa Bauer, Nicolas Papageorgiou, Francois Ferron, Tim Donselaar, Arno L.W. van Vliet, Tatiana M Shamorkina, Joost Snijder, Bruno Canard, Etienne Decroly, Andrea Brancale, Tzviya Zeev-Ben-Mordehai, Friedrich Förster, Frank J.M. van Kuppeveld, Bruno Coutard
Science Advances 8 (2022)10.1126/sciadv.abj7615
1H, 13C, 15N backbone resonance assignment of apo and ADP-ribose bound forms of the macro domain of Hepatitis E virus through solution NMR spectroscopy
Maria Politi, Angelo Gallo, Georgios Bouras, Maria Birkou, Bruno Canard, Bruno Coutard, Georgios Spyroulias
Biomolecular NMR Assignments (2022)10.1007/s12104-022-10111-5
Facile access to 4′-( N -acylsulfonamide) modified nucleosides and evaluation of their inhibitory activity against SARS-CoV-2 RNA cap N 7-guanine-methyltransferase nsp14
Romain Amador, Adrien Delpal, Bruno Canard, Jean-Jacques Vasseur, Etienne Decroly, Françoise Debart, Guillaume Clavé, Michael Smietana
Organic & Biomolecular Chemistry 20:7582-7586 (2022)10.1039/d2ob01569b
Synthesis, Structure–Activity Relationships, and Antiviral Profiling of 1-Heteroaryl-2-Alkoxyphenyl Analogs As Inhibitors of SARS-CoV-2 Replication
Dorothée Bardiot, Laura Vangeel, Mohamed Koukni, Philippe Arzel, Marleen Zwaagstra, Heyrhyoung Lyoo, Patrick Wanningen, Shamshad Ahmad, Linlin Zhang, Xinyuanyuan Sun, Adrien Delpal, Cecilia Eydoux, Jean-Claude Guillemot, Eveline Lescrinier, Hugo Klaassen, Pieter Leyssen, Dirk Jochmans, Karolien Castermans, Rolf Hilgenfeld, Colin Robinson, Etienne Decroly, Bruno Canard, Eric J Snijder, Martijn J Van Hemert, Frank Van van Kuppeveld, Patrick Chaltin, Johan Neyts, Steven De de Jonghe, Arnaud Marchand
Molecules 27:1052 (2022)10.3390/molecules27031052
Structure-function analysis of the nsp14 N7-guanine methyltransferase reveals an essential role in Betacoronavirus replication
Natacha S Ogando, Priscila El Kazzi, Jessika C Zevenhoven-Dobbe, Brenda W Bontes, Alice Decombe, Clara C S Posthuma, Volker Thiel, Bruno Canard, Francois Ferron, Etienne Decroly, Eric J Snijder
Proceedings of the National Academy of Sciences of the United States of America (2021)10.1101/2021.05.17.444407
System-oriented optimization of multi-target 2,6-diaminopurine derivatives: Easily accessible broad-spectrum antivirals active against flaviviruses, influenza virus and SARS-CoV-2
Ilaria Vicenti, Maria Grazia Martina, Adele Boccuto, Marta de Angelis, Giorgia Giavarini, Filippo Dragoni, Serena Marchi, Claudia Maria Trombetta, Emmanuele Crespan, Giovanni Maga, Cecilia Eydoux, Etienne Decroly, Emanuele Montomoli, Lucia Nencioni, Maurizio Zazzi, Marco Radi
European Journal of Medicinal Chemistry 224:113683 (2021)10.1016/j.ejmech.2021.113683
Fragment-based drug design targeting syntenin PDZ2 domain involved in exosomal release and tumour spread
Manon Garcia, Laurent Hoffer, Raphaël Leblanc, Fatiha Benmansour, Mikael Feracci, Carine Derviaux, Antonio Luis Egea-Jimenez, Philippe Roche, Pascale Zimmermann, Xavier Morelli, Karine Barral
European Journal of Medicinal Chemistry 223:113601 (2021)10.1016/j.ejmech.2021.113601
Fluoxetine targets an allosteric site in the enterovirus 2C AAA+ ATPase and stabilizes the hexameric complex
Daniel L Hurdiss, Priscila El Kazzi, Lisa Bauer, Nicolas Papageorgiou, Francois Ferron, Tim Donselaar, Arno L W van Vliet, Bruno Canard, Etienne Decroly, Andrea Brancale, Tzviya Zeev-Ben-Mordehai, Friedrich Förster, Frank J M van Kuppeveld, Bruno Coutard
A high-throughput fluorescence polarization assay to discover inhibitors of arenavirus and coronavirus exoribonucleases
Sergio Hernández, Mikael Feracci, Carolina Trajano de Jesus, Priscila El-Kazzi, Rafik Kaci, Laura Garlatti, Etienne Decroly, Bruno Canard, Francois Ferron, Karine Alvarez
An appeal for an objective, open, and transparent scientific debate about the origin of SARS-CoV-2
Jacques van Helden, Colin Butler, Guillaume Achaz, Bruno Canard, Didier Casane, Jean-Michel Claverie, Fabien Colombo, Virginie Courtier-Orgogozo, Richard Ebright, François Graner, Milton Leitenberg, Serge Morand, Nikolai Petrovsky, Rossana Segreto, Etienne Decroly, José Halloy
The Lancet 398:1402-1404 (2021)10.1016/S0140-6736(21)02019-5
Structure and Sequence Requirements for RNA Capping at the Venezuelan Equine Encephalitis Virus RNA 5′ End
Oney Ortega Granda, Coralie Valle, Ashleigh Shannon, Etienne Decroly, Bruno Canard, Bruno Coutard, Nadia Rabah
Journal of Virology 95 (2021)10.1128/JVI.00777-21
The enzymes for genome size increase and maintenance of large (+)RNA viruses
Francois Ferron, Bhawna Sama, Etienne Decroly, Bruno Canard
Trends in Biochemical Sciences (2021)10.1016/j.tibs.2021.05.006
Implication des Protéine Convertase humaine sur l'activation de la Spike protein du Sars-CoV-2
Adrien Delpal, Priscila Sutto-Ortiz, Bruno Coutard, Étienne Decroly, Nabil G Seidah
(2021)
Analyse structurale de la nouvelle N7 guanine-MTase du virus de la Brème bordelière
Pierre Gauffre, Ashleigh Shannon, Bhawna Sama, Bruno Canard, François Ferron
25:68-74 (2021)10.1684/vir.2021.0892
Tracing the origins of SARS-COV-2 in coronavirus phylogenies: a review
Erwan Sallard, José Halloy, Didier Casane, Etienne Decroly, Jacques van Helden
Environmental Chemistry Letters 19:769-785 (2021)10.1007/s10311-020-01151-1
Observation of arenavirus nucleoprotein heptamer assembly
Nicolas Papageorgiou, Afroditi Vaitsopoulou, Awa Diop, Thi Hong van Nguyen, Bruno Canard, Karine Alvarez, Francois Ferron
FEBS Open Bio (2021)10.1002/2211-5463.13106
A fluorescence-based high throughput-screening assay for the SARS-CoV RNA synthesis complex
Cecilia Eydoux, Veronique Fattorini, Ashleigh Shannon, Thi-Tuyet-Nhung Le, Bruno Didier, Bruno Canard, Jean-Claude Guillemot
Journal of Virological Methods 288:114013 (2021)10.1016/j.jviromet.2020.114013
Dengue Virus NS5 Transcribes Metabolite-Capped, RIG-I Sensitive vRNAs
Brandon Schweibenz, Mihai Solotchi, Etienne Decroly, Barbara Selisko, Bruno Canard, Smita Patel
Biophysical Journal 120:135a (2021)10.1016/j.bpj.2020.11.1017
COVID-19 epidemiologic surveillance using wastewater
Virender Sharma, Chetan Jinadatha, Eric Lichtfouse, Etienne Decroly, Jacques van Helden, Hosoon Choi, Piyali Chatterjee
Environmental Chemistry Letters 19:1911-1915 (2021)10.1007/s10311-021-01188-w
Simeprevir Potently Suppresses SARS-CoV-2 Replication and Synergizes with Remdesivir
Ho Sing Lo, Kenrie Pui Yan Hui, Hei-Ming Lai, Xu He, Khadija Shahed Khan, Simranjeet Kaur, Junzhe Huang, Zhongqi Li, Anthony Chan, Hayley Hei-Yin Cheung, Ka-Chun Ng, John Chi Wang Ho, Yu Wai Chen, Bowen Ma, Peter Man-Hin Cheung, Donghyuk Shin, Kaidao Wang, Meng-Hsuan Lee, Cécilia Eydoux, Cecilia Eydoux, Jean-Claude Guillemot, Bruno Canard, Kuen-Phon Wu, Po-Huang Liang, Ivan Dikic, Zhong Zuo, Francis Chan, David Hui, Vincent Mok, Kam-Bo Wong, Chris Ka Pun Mok, Ho Ko, Wei Shen Aik, Michael Chi Wai Chan, Wai-Lung Ng
ACS Central Science 7:792-802 (2021)10.1021/acscentsci.0c01186
The methyltransferase domain of the Respiratory Syncytial Virus L protein catalyzes cap N7 and 2’-O-methylation
Priscila Sutto-Ortiz, Sergey Tcherniuk, Nina Ysebaert, Pravien Abeywickrema, Mathieu Noël, Alice Decombe, Françoise Debart, Jean-Jacques Vasseur, Bruno Canard, Dirk Roymans, Peter Rigaux, Jean-François Eléouët, Etienne Decroly
PLoS Pathogens 17:1-20 (2021)10.1371/journal.ppat.1009562
Efficient access to 3′-deoxy-3′-(4-substituted-1,2,3-triazol-1-yl)-thymidine derivatives via ligand-promoted CuAAC
Laura Garlatti, Raphaël Huet, Karine Alvarez
Tetrahedron 92:132252 (2021)10.1016/j.tet.2021.132252
The WHO mission report struggles to trace the origins of the SARS-CoV-2 epidemic
Etienne Decroly, Jean-Michel Claverie, Bruno Canard
Virologie 1-7 (2021)
Combining Antivirals and Immunomodulators to Fight COVID-19
Vincent Feuillet, Bruno Canard, Alain Trautmann
Trends in Immunology 42:31-44 (2021)10.1016/j.it.2020.11.003
First insights into the structural features of Ebola virus methyltransferase activities
Coralie Valle, Baptiste Martin, François Ferron, Véronique Roig-Zamboni, Aline Desmyter, Françoise Debart, Jean Jacques Vasseur, Bruno Canard, Bruno Coutard, Etienne Decroly
Nucleic Acids Research 49:1737-1748 (2021)10.1093/nar/gkaa1276
Favipiravir strikes the SARS-CoV-2 at its Achilles heel, the RNA polymerase
A. Shannon, B. Selisko, Ntt Le, J. Huchting, F. Touret, G. Piorkowski, V. Fattorini, F. Ferron, Etienne Decroly, C Meier, B. Coutard, O. Peersen, B. Canard
Tracing the origins of SARS-CoV-2 in coronavirus phylogenies
Erwan Sallard, José Halloy, Didier Casane, Etienne Decroly, Jacques van Helden
Médecine/Sciences 36 (2020)10.1051/medsci/2020123
Pharmacological inhibition of syntenin PDZ2 domain impairs breast cancer cell activities and exosome loading with syndecan and EpCAM cargo
R. Leblanc, R. Kashyap, K. Barral, A.L. Egea‐jimenez, D. Kovalskyy, M. Feracci, M. Garcia, C. Derviaux, S. Betzi, R. Ghossoub, M. Platonov, P. Roche, X. Morelli, L. Hoffer, Pascale Zimmermann
Journal of Extracellular Vesicles 10 (2020)10.1002/jev2.12039
Targeting furin activity through in silico and in vitro drug repurposing strategy for SARS-CoV-2 spike glycoprotein cleavage repression
Bruno Villoutreix, John Creemers, Yannick Léger, Geraldine Siegfried, Etienne Decroly, Serge Evrard, Abdel-Majid Khatib
Drugs against SARS‐CoV ‐2: What do we know about their mode of action?
Coralie Valle, Baptiste Martin, Franck Touret, Ashleigh Shannon, Bruno Canard, Jean‐claude Guillemot, Bruno Coutard, Etienne Decroly
Reviews in Medical Virology 30:1-10 (2020)10.1002/rmv.2143
Synthesis and biological evaluation of novel flexible nucleoside analogues that inhibit flavivirus replication in vitro
Joy Thames, Charles Waters, Coralie Valle, Marcella Bassetto, Wahiba Aouadi, Baptiste Martin, Barbara Selisko, Arissa Falat, Bruno Coutard, Andrea Brancale, Bruno Canard, Etienne Decroly, Katherine Seley-Radtke
Bioorganic and Medicinal Chemistry Letters 28:115713 (2020)10.1016/j.bmc.2020.115713
Mutations on VEEV nsP1 relate RNA capping efficiency to ribavirin susceptibility
Nadia Rabah, Oney Ortega Granda, Gilles Quérat, Bruno Canard, Etienne Decroly, Bruno Coutard
Antiviral Research 182:104883 (2020)10.1016/j.antiviral.2020.104883
Structure-based drug repositioning over the human TMPRSS2 protease domain: search for chemical probes able to repress SARS-CoV-2 Spike protein cleavages
Natesh Singh, Etienne Decroly, Abdel-Majid Khatib, Bruno Villoutreix
European Journal of Pharmaceutical Sciences 153:105495 (2020)10.1016/j.ejps.2020.105495
Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis
Ashleigh Shannon, Barbara Selisko, Nhung-Thi-Tuyet Le, Johanna Huchting, Franck Touret, Géraldine Piorkowski, Véronique Fattorini, Francois Ferron, Etienne Decroly, Chris Meier, Bruno Coutard, Olve Peersen, Bruno Canard
Nature Communications 11:4682 (2020)10.1038/s41467-020-18463-z
Snapshots of ADp-ribose bound to Getah virus macro domain reveal an intriguing choreography
Ana Sofia Ferreira-Ramos, Gerlind Sulzenbacher, Bruno Canard, Bruno Coutard
Scientific Reports 10 (2020)10.1038/s41598-020-70870-w
Synthesis of adenine dinucleosides SAM analogs as specific inhibitors of SARS-CoV nsp14 RNA cap guanine-N7-methyltransferase
Rostom Ahmed‐belkacem, Priscila Sutto-Ortiz, Mathis Guiraud, Bruno Canard, Jean-Jacques Vasseur, Etienne Decroly, Francoise Debart
European Journal of Medicinal Chemistry 201:112557 (2020)10.1016/j.ejmech.2020.112557
In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication
Franck Touret, Magali Gilles, Karine Barral, Antoine Nougairède, Jacques van Helden, Etienne Decroly, Xavier de Lamballerie, Bruno Coutard
Scientific Reports 10:13093 (2020)10.1038/s41598-020-70143-6
Obesity of mice lacking VAP-1/SSAO by Aoc3 gene deletion is reproduced in mice expressing a mutated vascular adhesion protein-1 (VAP-1) devoid of amine oxidase activity
Valentin Jargaud, Sandy Bour, François Tercé, Xavier Collet, Philippe Valet, Anne Bouloumié, Jean-Claude Guillemot, Pascale Mauriège, Sirpa Jalkanen, Craig Stolen, Marko Salmi, David Smith, Christian Carpéné
Journal of Physiology and Biochemistry 77:141-154 (2020)10.1007/s13105-020-00756-y
Brothers in Arms: Structure, Assembly and Function of Arenaviridae Nucleoprotein
Nicolas Papageorgiou, Maria Spiliopoulou, Thi-Hong van Nguyen, Afroditi Vaitsopoulou, Elsie Yekwa Laban, Karine Alvarez, Irene Margiolaki, Bruno Canard, Francois Ferron
Viruses 12:772 (2020)10.3390/v12070772
Novel Class of Chikungunya Virus Small Molecule Inhibitors That Targets the Viral Capping Machinery
Rana Abdelnabi, Kristina Kovacikova, Julia Moesslacher, Kim Donckers, Verena Battisti, Pieter Leyssen, Thierry Langer, Gerhard Puerstinger, Gilles Quérat, Changqing Li, Etienne Decroly, Ali Tas, Arnaud Marchand, Patrick Chaltin, Bruno Coutard, Martijn van Hemert, Johan Neyts, Leen Delang
Antimicrobial Agents and Chemotherapy 64 (2020)10.1128/AAC.00649-20
To something, Covid is good: SFV 2.0
Grégory Caignard, Etienne Decroly, Maria Dimitrova, David Gilmer, Anne-Sophie Gosselin-Grenet, Henri Gruffat, Sandra Martin-Latil, Pascale Massin, Sandie Munier, Mylène Ogliastro, Aure Saulnier, Noël Tordo
Virologie 24:133-134 (2020)10.1684/vir.2020.0846
Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites
Ashleigh Shannon, Nhung Thi-Tuyet Le, Barbara Selisko, Cecilia Eydoux, Karine Alvarez, Jean-Claude Guillemot, Etienne Decroly, Olve Peersen, Francois Ferron, Bruno Canard
Antiviral Research 178:104793 (2020)10.1016/j.antiviral.2020.104793
Discovery, SAR study and ADME properties of methyl 4-amino-3-cyano-1-(2-benzyloxyphenyl)-1 H -pyrazole-5-carboxylate as an HIV-1 replication inhibitor
Jeanne Fichez, Cathia Soulié, Laurent Le Corre, Sophie Sayon, Stéphane Priet, Karine Alvarez, Olivier Delelis, Patrick Gizzi, Guillaume Prestat, Christine Gravier-Pelletier, Anne-Geneviève Marcelin, Vincent Calvez, Patricia Busca
RSC Medicinal Chemistry 11:577-582 (2020)10.1039/D0MD00025F
Incretin combination therapy for the treatment of non-alcoholic steatohepatitis
Aimo Kannt, Andreas Nygaard Madsen, Claire Kammermeier, Ralf Elvert, Tim Klöckener, Martin Bossart, Torsten Haack, Andreas Evers, Katrin Lorenz, W. Hennerici, Corinne Rocher, Zsolt Böcskei, Jean-Claude Guillemot, Vincent Mikol, François Pattou, Bart Staels, Michael Wagner
Diabetes, Obesity and Metabolism Online ahead of print. (2020)10.1111/dom.14035
A N7-guanine RNA cap methyltransferase signature-sequence as a genetic marker of large genome, non-mammalian Tobaniviridae
Francois Ferron, Humberto Julio Debat, Ashleigh Shannon, Etienne Decroly, Bruno Canard
NAR Genomics and Bioinformatics 2 (2020)10.1093/nargab/lqz022
Design, Synthesis and Discovery of N,N’ ‐Carbazoyl‐aryl‐urea Inhibitors of Zika NS5 Methyltransferase and Virus Replication
Sharon Spizzichino, Giulio Mattedi, Kate Lauder, Coralie Valle, Wahiba Aouadi, Bruno Canard, Etienne Decroly, Suzanne Kaptein, Johan Neyts, Carl Graham, Zakary Sule, David Barlow, Romano Silvestri, Daniele Castagnolo
ChemMedChem 15:385-390 (2020)10.1002/cmdc.201900533
The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade
Bruno Coutard, Coralie Valle, Xavier de Lamballerie, Bruno Canard, Nabil G Seidah, Etienne Decroly
Antiviral Research 176:104742 (2020)10.1016/j.antiviral.2020.104742
Carneic Acids from an Endophytic Phomopsis sp. as Dengue Virus Polymerase Inhibitors
Laure-Anne Peyrat, Véronique Eparvier, Cecilia Eydoux, Jean-Claude Guillemot, Marc Litaudon, Didier Stien
Journal of Natural Products 83:2330-2336 (2020)10.1021/acs.jnatprod.9b01169
In silico molecular target prediction unveils mebendazole as a potent MAPK14 inhibitor
Jeremy Ariey‐bonnet, Kendall Carrasco, Marion Le Grand, Laurent Hoffer, Stéphane Betzi, Mikael Feracci, Philipp A Tsvetkov, François Devred, Yves Collette, Xavier Morelli, Pedro Ballester, Eddy Pasquier
Molecular Oncology 14:3083-3099 (2020)10.1002/1878-0261.12810
The C-Terminal Domain of the Sudan Ebolavirus L Protein Is Essential for RNA Binding and Methylation
Coralie Valle, Baptiste Martin, Francoise Debart, Jean-Jacques Vasseur, Isabelle Imbert, Bruno Canard, Bruno Coutard, Etienne Decroly
Journal of Virology 94:e00520-20 (2020)10.1128/JVI.00520-20
Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6
Olivier Fuchsbauer, Paolo Swuec, Claire Zimberger, Beatrice Amigues, Sébastien Levesque, Daniel Agudelo, Alexis Duringer, Antonio Chaves-Sanjuan, Silvia Spinelli, Geneviève Rousseau, Minja Velimirovic, Martino Bolognesi, Alain Roussel, Christian Cambillau, Sylvain Moineau, Yannick Doyon, Adeline Goulet
Molecular Cell 76:922-937.e7 (2019)10.1016/j.molcel.2019.09.012
Arenaviridae exoribonuclease presents genomic RNA edition capacity.
Elsie Laban Yekwa, Chutima Aphibanthammakit, Xavier Carnec, Caroline Picard, Bruno Canard, Sylvain Baize, Francois Ferron
(2019)10.1101/541698
Phase transition and amyloid formation by a viral protein as an additional molecular mechanism of virus-induced cell toxicity
Edoardo Salladini, Claire Debarnot, Vincent Delauzun, Maria Grazia Murrali, Priscila Sutto-Ortiz, Silvia Spinelli, Roberta Pierattelli, Christophe Bignon, Sonia Longhi
(2019)
Structure of the Respiratory Syncytial Virus Polymerase Complex
Morgan S.A. Gilman, Cheng Liu, Amy Fung, Ishani Behera, Paul Jordan, Peter Rigaux, Nina Ysebaert, Sergey Tcherniuk, Julien Sourimant, Jean-François Éléouët, Priscila Sutto-Ortiz, Etienne Decroly, Dirk Roymans, Zhinan Jin, Jason S Mclellan
Cell 179:193-204.e14 (2019)10.1016/j.cell.2019.08.014
The Curious Case of the Nidovirus Exoribonuclease: Its Role in RNA Synthesis and Replication Fidelity
Natacha S Ogando, Francois Ferron, Etienne Decroly, Bruno Canard, Clara C Posthuma, Eric J Snijder
Frontiers in Microbiology 10 (2019)10.3389/fmicb.2019.01813
A61 Large RNA genomes: Is RNA polymerase fidelity enough?
F. Ferron, B. Canard
Virus Evolution 5:179-184 (2019)10.1093/ve/vez002.060
Identification of a Nidovirales Orf1a N7-guanine cap Methyltransferase signature- sequence as a genetic marker of large genome Tobaniviridae Running title: RNA cap N7-guanine Methyltransferase in Tobaniviridae Orf1a
Francois Ferron, Humberto Julio Debat, Etienne Decroly, Bruno Canard
(2019)10.1101/639369
Deciphering the Nucleotide and RNA Binding Selectivity of the Mayaro Virus Macro Domain
Aikaterini Tsika, Efstathios Melekis, Sofia-Antigoni Tsatsouli, Nicolas Papageorgiou, Maria Mate, Bruno Canard, Bruno Coutard, Detlef Bentrop, Georgios Spyroulias
Journal of Molecular Biology 431:2283-2297 (2019)10.1016/j.jmb.2019.04.013
Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose
Garyfallia Makrynitsa, Dioni Ntonti, Konstantinos Marousis, Maria Birkou, Minos-Timotheos Matsoukas, Sam Asami, Detlef Bentrop, Nicolas Papageorgiou, Bruno Canard, Bruno Coutard, Georgios Spyroulias
Journal of Structural Biology 206:119-127 (2019)10.1016/j.jsb.2019.02.008
Gln151 of HIV-1 reverse transcriptase acts as a steric gate towards clinically relevant acyclic phosphonate nucleotide analogues.
Antoine Frangeul, Cécile Bussetta, Jérôme Deval, Karine Barral, Karine Alvarez, Bruno Canard
Antiviral Therapy 13:115-24 (2019)
Comparative study of chikungunya Virus-Like Particles and Pseudotyped-Particles used for serological detection of specific immunoglobulin M
Gérald Theillet, Jérôme Martinez, Christophe Steinbrugger, Dimitri Lavillette, Bruno Coutard, Nicolas Papageorgiou, Pascal Dalbon, Isabelle Leparc-Goffart, Frederic Bedin
Virology 529:195-204 (2019)10.1016/j.virol.2019.01.027
C3P3-G1: first generation of a eukaryotic artificial cytoplasmic expression system
Philippe H Jaïs, Etienne Decroly, Eric Jacquet, Marine Le boulch, Aurélien Jaïs, Olivier Jean-Jean, Heather Eaton, Prishila Ponien, Frédérique Verdier, Bruno Canard, Sergio Gonçalves, Stéphane Chiron, Maude Le Gall, Patrick Mayeux, Maya Shmulevitz
Nucleic Acids Research 47:2681-2698 (2019)10.1093/nar/gkz069
Metal chelators for the inhibition of the lymphocytic choriomeningitis virus endonuclease domain
Magali Saez-Ayala, Elsie Laban Yekwa, Clémence Mondielli, Loic Roux, Sergio Hernández, Fabrice Bailly, Philippe Cotelle, Dominga Rogolino, Bruno Canard, Francois Ferron, Karine Alvarez
Antiviral Research 162:79-89 (2019)10.1016/j.antiviral.2018.12.008
Antiviral Compounds from Codiaeum peltatum Targeted by a Multi-informative Molecular Networks Approach
Florent Olivon, Simon Remy, Gwendal Grelier, Cécile Apel, Cecilia Eydoux, Jean-Claude Guillemot, Johan Neyts, Leen Delang, David Touboul, Fanny Roussi, Marc Litaudon
Journal of Natural Products 82:330-340 (2019)10.1021/acs.jnatprod.8b00800
Approved drugs screening against the nsP1 capping enzyme of Venezuelan equine encephalitis virus using an immuno-based assay
Ana Ferreira-Ramos, Changqing Li, Cecilia Eydoux, Jean Marie Contreras, Christophe Morice, Gilles Querat, Alba Gigante, María-Jesús Pérez Pérez, Marie-Louise Jung, Bruno Canard, Jean-Claude Guillemot, Etienne Decroly, Bruno Coutard
Antiviral Research 163:59-69 (2019)10.1016/j.antiviral.2019.01.003
Identification of a new natural gastric lipase inhibitor from star anise
Jannet Kamoun, Renaud Rahier, Mohamed Sellami, Imed Koubaa, Pascal Mansuelle, Régine Lebrun, Alexandra Berlioz-Barbier, Michele Fiore, Karine Alvarez, Abdelkarim Abousalham, Frédéric Carrière, Ahmed Aloulou
Food and Function 10:469-478 (2019)10.1039/c8fo02009d
FTSJ3 is an RNA 2′-O-methyltransferase recruited by HIV to avoid innate immune sensing
Mathieu Ringeard, Virginie Marchand, Etienne Decroly, Yuri Motorin, Yamina Bennasser
Nature 565:500-504 (2019)10.1038/s41586-018-0841-4
Structure and oligomerization state of the C-terminal region of the Middle East respiratory syndrome coronavirus nucleoprotein
Thi Hong Van Nguyen, Julie Lichiere, Bruno Canard, Nicolas Papageorgiou, Sarah Attoumani, Francois Ferron, Bruno Coutard
Acta crystallographica Section D : Structural biology [1993-...] 75:8-15 (2019)10.1107/S2059798318014948
Synthesis of Adenine Dinucleosides 2′,5′-Bridged by Sulfur-Containing Linkers as Bisubstrate SAM Analogues for Viral RNA 2′- O -Methyltransferases
Rostom Ahmed‐belkacem, Priscila Sutto Ortiz, Etienne Decroly, Jean-Jacques Vasseur, Françoise Debart
European Journal of Organic Chemistry 2019 (2019)10.1002/ejoc.201901120
Optimization of a fragment linking hit toward Dengue and Zika virus NS5 methyltransferases inhibitors
Jessica Hernandez, Laurent Hoffer, Bruno Coutard, Gilles Quérat, Philippe Roche, Xavier Morelli, Etienne Decroly, Karine Barral
European Journal of Medicinal Chemistry 161:323-333 (2019)10.1016/j.ejmech.2018.09.056
Ebola virus L protein harbors a new enzymatic activity involved in the internal methylation of RNAs
Baptiste Martin, Coralie Valle, Bruno Coutard, Bruno Canard, Françoise Debart, Etienne Decroly
Médecine/Sciences 34:919-921 (2018)10.1051/medsci/2018230
The methyltransferase domain of the Sudan ebolavirus L protein specifically targets internal adenosines of RNA substrates, in addition to the cap structure
Baptiste Martin, Bruno Coutard, Théo Guez, Guido Paesen, Bruno Canard, Françoise Debart, Jean-Jacques Vasseur, Jonathan Grimes, Etienne Decroly
Nucleic Acids Research 46:7902-7912 (2018)10.1093/nar/gky637
Galactolipase activity of Talaromyces thermophilus lipase on galactolipid micelles, monomolecular films and UV-absorbing surface-coated substrate
Inès Belhaj, Sawsan Amara, Goetz Parsiegla, Priscila Sutto-Ortiz, Moulay Sahaka, Hafedh Belghith, Audric Rousset, Dominique Lafont, Frédéric Carriere
Biochimica et Biophysica Acta Molecular and Cell Biology of Lipids 1863:1006-1015 (2018)10.1016/j.bbalip.2018.05.016
A Continuous and Sensitive Spectrophotometric Assay for Lipase and Phospholipase A Activities Using α-Eleostearic Acid-Containing Substrates
Meddy El Alaoui, Laurent Soulère, Alexandre Noiriel, Priscila Sutto-Ortiz, Lucie Grand, Florence Popowycz, Jorge Alberto Rodríguez-González, Yves Queneau, Abdelkarim Abousalham
Methods in Molecular Biology 1835:119-128 (2018)10.1007/978-1-4939-8672-9_5
Innate immunity evasion mechanisms of filoviruses
Baptiste Martin, Etienne Decroly
Med Sci (Paris) 34:671-677 (2018)10.1051/medsci/20183408013
A Vaccine Platform against Arenaviruses Based on a Recombinant Hyperattenuated Mopeia Virus Expressing Heterologous Glycoproteins
Xavier Carnec, Mathieu Mateo, Audrey Page, Stéphanie Reynard, Jimmy Hortion, Caroline Picard, Elsie Yekwa, Laura Barrot, Stéphane Barron, Audrey Vallve, Hervé Raoul, Caroline Carbonnelle, Francois Ferron, Sylvain Baize
Journal of Virology 92:e02230-17 (2018)10.1128/JVI.02230-17
Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors
Ludovic Halby, Nils Marechal, Dany Pechalrieu, Vincent Cura, Don-Marc Franchini, Céline Faux, Frédéric Alby, Nathalie Troffer-Charlier, Srikanth Kudithipudi, Albert Jeltsch, Wahiba Aouadi, Etienne Decroly, Jean-Claude Guillemot, Patrick Page, Clotilde Ferroud, Luc Bonnefond, Dominique Guianvarc’h, Jean Cavarelli, Paola Arimondo
Philosophical Transactions of the Royal Society B: Biological Sciences 373:20170072 (2018)10.1098/rstb.2017.0072
IR spectroscopy analysis of pancreatic lipase-related protein 2 interaction with phospholipids: 2. Discriminative recognition of various micellar systems and characterization of PLRP2-DPPC-bile salt complexes
Eduardo Mateos-Diaz, Priscila Sutto-Ortiz, Moulay Sahaka, Deborah Byrne, Hélène Gaussier, Frédéric Carrière
Chemistry and Physics of Lipids 211:66-76 (2018)10.1016/j.chemphyslip.2017.11.012
IR spectroscopy analysis of pancreatic lipase-related protein 2 interaction with phospholipids: 3. Monitoring DPPC lipolysis in mixed micelles
Eduardo Mateos-Diaz, Priscila Sutto-Ortiz, Moulay Sahaka, Jorge Rodriguez, Frédéric Carrière
Chemistry and Physics of Lipids 211:77-85 (2018)10.1016/j.chemphyslip.2017.11.009
Crystal structures of Lymphocytic choriomeningitis virus endonuclease domain complexed with diketo-acid ligands
Magali Saez-Ayala, Elsie Laban Yekwa, Mauro Carcelli, Bruno Canard, Karine Alvarez, Francois Ferron
International Union of Crystallography journal 5:223-235 (2018)10.1107/S2052252518001021
Crystal structures ofLymphocytic choriomeningitis virusendonuclease domain complexed with diketo-acid ligands
Magali Saez-Ayala, Elsie Yekwa, Mauro Carcelli, Bruno Canard, Karine Alvarez, Francois Ferron
International Union of Crystallography journal 5:223-235 (2018)10.1107/s2052252518001021
Structural and Functional Basis of the Fidelity of Nucleotide Selection by Flavivirus RNA-Dependent RNA Polymerases
Barbara Selisko, Nicolas Papageorgiou, Francois Ferron, Bruno Canard
Viruses 10:59 (2018)10.3390/v10020059
Filovirus proteins for antiviral drug discovery: Structure/function of proteins involved in assembly and budding
Baptiste Martin, Olivier Reynard, Viktor Volchkov, Etienne Decroly
Antiviral Research 150:183-192 (2018)10.1016/j.antiviral.2017.12.022
Synthesis and substrate properties towards HIV-1 reverse transcriptase of new diphosphate analogues of 9-[(2-phosphonomethoxy)ethyl]adenine
Wolfgang Hg Laux, Stephane Priet, Karine Alvarez, Suzanne Peyrottes, Christian Perigaud
Antiviral Chemistry and Chemotherapy 26:204020661875763 (2018)10.1177/2040206618757636
Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA
Francois Ferron, Lorenzo Subissi, Ana Theresa Silveira de Morais, Nhung Thi Tuyet Le, Marion Sevajol, Laure Gluais, Etienne Decroly, Clemens Vonrhein, Gérard Bricogne, Bruno Canard, Isabelle Imbert
Proceedings of the National Academy of Sciences of the United States of America 115:E162-E171 (2018)10.1073/pnas.1718806115
A Continuous and Sensitive Spectrophotometric Assay for Lipase and Phospholipase A Activities using α-Eleostearic Acid-containing Substrates
Meddy El Alaoui, Laurent Soulère, Alexandre Noiriel, Priscila Sutto-Ortiz, Lucie Grand, Florence Popowycz, Jorge Alberto Rodríguez-González, Yves Queneau, Abdelkarim Abousalham
119-128 (2018)
How to Control HTLV-1-Associated Diseases: Preventing de Novo Cellular Infection Using Antiviral Therapy
Amandine Pasquier, Sandrine Alais, Loic Roux, Marie-Isabelle Thoulouze, Karine Alvarez, Chloé Journo, Hélène Dutartre, Renaud Mahieux
Frontiers in Microbiology 9:278 (2018)10.3389/fmicb.2018.00278
Toscana virus nucleoprotein oligomer organization observed in solution
Amal Baklouti, Adeline Goulet, Julie Lichiere, Bruno Canard, Remi Charrel, Francois Ferron, Bruno Coutard, Nicolas Papageorgiou
Acta crystallographica Section D : Structural biology [1993-...] 73:650-659 (2017)10.1107/S2059798317008774
NF-L in cerebrospinal fluid and serum is a biomarker of neuronal damage in an inducible mouse model of neurodegeneration
Anthony Brureau, Véronique Blanchard-Bregeon, Catherine Pech, Stéphanie Hamon, Pascal Chaillou, Jean-Claude Guillemot, Pascal Barneoud, Philippe Bertrand, Laurent Pradier, Thomas Rooney, Nathalie Schussler
Neurobiology of Disease 104:73-84 (2017)10.1016/j.nbd.2017.04.007
Toward the identification of viral cap-methyltransferase inhibitors by fluorescence screening assay
Wahiba Aouadi, C Ecilia Eydoux, Bruno Coutard, Baptiste Martin, Françoise Debart, Jean-Jacques Vasseur, Jean Marie Contreras, Christophe Morice, Gilles Quérat, Marie-Louise Jung, Bruno Canard, Jean-Claude Guillemot, Etienne Decroly
Antiviral Research 144:330-339 (2017)10.1016/j.antiviral.2017.06.021
Activity inhibition and crystal polymorphism induced by active-site metal swapping
Elsie Yekwa, Joelle Khourieh, Bruno Canard, Nicolas Papageorgiou, Francois Ferron
Acta crystallographica Section D : Structural biology [1993-...] 73:641-649 (2017)10.1107/S205979831700866X
Understanding the Mechanism of the Broad-Spectrum Antiviral Activity of Favipiravir (T-705): Key Role of the F1 Motif of the Viral Polymerase
Rana Abdelnabi, Ana Theresa Silveira De Morais, Pieter Leyssen, Isabelle Imbert, Stéphanie Beaucourt, Hervé Blanc, Mathy Froeyen, Marco Vignuzzi, Bruno Canard, Johan Neyts, Leen Delang
Journal of Virology 91:e00487-17 (2017)10.1128/JVI.00487-17
Biochemical principles and inhibitors to interfere with viral capping pathways
Etienne Decroly, Bruno Canard
Current Opinion in Virology 24:87-96 (2017)10.1016/j.coviro.2017.04.003
Filovirus proteins for antiviral drug discovery: Structure/function bases of the replication cycle
Baptiste Martin, Bruno Canard, Etienne Decroly
Antiviral Research 141:48-61 (2017)10.1016/j.antiviral.2017.02.004
Binding site density enables paralog-specific activity of SLM2 and Sam68 proteins in Neurexin2 AS4 splicing control
Marina Danilenko, Caroline Dalgliesh, Vittoria Pagliarini, Chiara Naro, Ingrid Ehrmann, Mikael Feracci, Mahsa Kheirollahi-Chadegani, Alison Tyson-Capper, Gavin Clowry, Philippe Fort, Cyril Dominguez, Claudio Sette, David Elliott
Nucleic Acids Research 45:gkw1277 (2017)10.1093/nar/gkw1277
Transcription and replication mechanisms of Bunyaviridae and Arenaviridae L proteins
Francois Ferron, Friedemann Weber, Juan de La Torre, Juan Reguera
Virus Research 234:118-134 (2017)10.1016/j.virusres.2017.01.018
Substrate selectivity of Dengue and Zika virus NS5 polymerase towards 2′-modified nucleotide analogues
Supanee Potisopon, Francois Ferron, Véronique Fattorini, Barbara Selisko, Bruno Canard
Antiviral Research 140:25-36 (2017)10.1016/j.antiviral.2016.12.021
Contribution to PNA-RNA Chimera Synthesis: One-Pot Microwave-Assisted Ugi Reaction to Obtain Dimeric Building Blocks
Reuben Ovadia, Clémence Mondielli, Jean-Jacques Vasseur, Carine Baraguey, Karine Alvarez
European Journal of Organic Chemistry 2017:469-475 (2017)10.1002/ejoc.201601190
Discovery of novel dengue virus NS5 methyltransferase non-nucleoside inhibitors by fragment-based drug design
Fatiha Benmansour, Iuni Trist, Bruno Coutard, Etienne Decroly, Gilles Querat, Andrea Brancale, Karine Barral
European Journal of Medicinal Chemistry 125:865-880 (2017)10.1016/j.ejmech.2016.10.007
Binding of the Methyl Donor S -Adenosyl-l-Methionine to Middle East Respiratory Syndrome Coronavirus 2′- O -Methyltransferase nsp16 Promotes Recruitment of the Allosteric Activator nsp10
Wahiba Aouadi, Alexandre Blanjoie, Jean-Jacques Vasseur, Françoise Debart, Bruno Canard, Etienne Decroly
Journal of Virology 91:1103-1119 (2017)10.1128/JVI.02217-16
Preparation of thiophenepropenamide derivatives as flavivirus inhibitors in treatment of viral infections
Karine Barral, Jean Claude Guillemot, Bruno Canard, Gilles Quérat, Karine Alvarez, Xavier de Lamballerie, Florence Mahuteau-Betzer, Cedric Poinsard
(2017)
Antiviral activity of [1,2,3]triazolo[4,5- d ]pyrimidin-7(6 H )-ones against chikungunya virus targeting the viral capping nsP1
Alba Gigante, Asier Gómez-Sanjuan, Leen Delang, Changqing Li, Oskía Bueno, Ana-María Gamo, Eva-María Priego, María-José Camarasa, Dirk Jochmans, Pieter Leyssen, Etienne Decroly, Bruno Coutard, Gilles Querat, Johan Neyts, María-Jesús Pérez-Pérez
Antiviral Research 144:216-222 (2017)10.1016/j.antiviral.2017.06.003
Zika Virus Methyltransferase: Structure and Functions for Drug Design Perspectives
Bruno Coutard, Karine Barral, Julie Lichiere, Barbara Selisko, Baptiste Martin, Wahiba Aouadi, Miguel Ortiz Lombardia, Françoise Debart, Jean-Jacques Vasseur, Jean Claude Guillemot, Bruno Canard, Etienne Decroly
Journal of Virology 91:2202-2218 (2017)10.1128/JVI.02202-16
Coxsackievirus B3 protease 3C: expression, purification, crystallization and preliminary structural insights
Stavroula Fili, Alexandros Valmas, Magdalini Christopoulou, Maria Spiliopoulou, Nikos Nikolopoulos, Julie Lichiere, Souzana Logotheti, Fotini Karavassili, Eleftheria Rosmaraki, Andrew Fitch, Jonathan Wright, Detlef Beckers, Thomas Degen, Gwilherm Nenert, Rolf Hilgenfeld, Nicolas Papageorgiou, Bruno Canard, Bruno Coutard, Irene Margiolaki
Acta crystallographica Section F : Structural biology communications [2014-...] 72:877-884 (2016)10.1107/S2053230X16018513
Filovirus proteins for antiviral drug discovery: A structure/function analysis of surface glycoproteins and virus entry
Baptiste Martin, Thomas Hoenen, Bruno Canard, Etienne Decroly
Antiviral Research 135:1-14 (2016)10.1016/j.antiviral.2016.09.001
Viral Macro Domains Reverse Protein ADP-Ribosylation
Changqing Li, Yannick Debing, Gytis Jankevicius, Johan Neyts, Ivan Ahel, Bruno Coutard, Bruno Canard
Journal of Virology 90:8478-8486 (2016)10.1128/JVI.00705-16
Betulinic Acid, The First Lupane-Type Triterpenoid Isolated from Both a Phomopsis sp. and Its Host Plant Diospyros carbonaria Benoist
Laure-Anne Peyrat, Véronique Eparvier, Cécilia Eydoux, Jean-Claude Guillemot, Marc Litaudon, Didier Stien
Chemistry and Biodiversity 14:e1600171 (2016)10.1002/cbdv.201600171
Protein-Protein Interaction Inhibition (2P21)-Oriented Chemical Library Accelerates Hit Discovery
Sabine Milhas, Brigitt Raux, Stéphane Betzi, Carine Derviaux, Philippe Roche, Audrey Restouin, Marie-Jeanne Basse, Etienne Rebuffet, Adrien Lugari, Marion Badol, Rudra Kashyap, Jean-Claude Lissitzky, Cecilia Eydoux, Veronique Hamon, Marie -Edith Gourde, Sebastien Combes, Pascale Zimmermann, Michel Aurrand-Lions, Thomas Roux, Catherine Rogers, Susanne Muller, Stefan Knapp, Eric Trinquet, Yves Collette, Jean-Claude Guillemot, Xavier Morelli
ACS Chemical Biology 11:2140-2148 (2016)10.1021/acschembio.6b00286
Screening of a Library of FDA-Approved Drugs Identifies Several Enterovirus Replication Inhibitors That Target Viral Protein 2C
Rachel Ulferts, S. Matthijn De Boer, Lonneke van Der Linden, Lisa Bauer, Hey Rhyoung Lyoo, Maria J. Mate, Julie Lichiere, Bruno Canard, Daphne Lelieveld, Wienand Omta, David Egan, Bruno Coutard, Frank J. M. Van Kuppeveld
Antimicrobial Agents and Chemotherapy 60:2627-2638 (2016)10.1128/AAC.02182-15
Predicting Conformational Disorder
Philippe Lieutaud, François Ferron, Sonia Longhi
1415:265-299 (2016)10.1007/978-1-4939-3572-7_14
The methyltransferase domain of dengue virus protein NS5 ensures efficient RNA synthesis initiation and elongation by the polymerase domain (vol 42, pg 11642, 2014)
Supanee Potisopon, Stephane Priet, Axelle Collet, Etienne Decroly, Bruno Canard, Barbara Selisko
Nucleic Acids Research 44:2974 (2016)10.1093/nar/gkv1294
Reevaluation of possible outcomes of infections with human immunodeficiency virus
C. Tamalet, Philippe Colson, Etienne Decroly, C. Dhiver, I. Ravaux, A. Stein, Didier Raoult
Clinical Microbiology and Infection 22:299-311 (2016)10.1016/j.cmi.2015.11.022
Novel 2-phenyl-5-[(E)-2-(thiophen-2-yl)ethenyl]-1,3,4-oxadiazole and 3-phenyl-5-[(E)-2-(thiophen-2-yl)ethenyl]-1,2,4-oxadiazole derivatives as dengue virus inhibitors targeting NS5 polymerase
Fatiha Benmansour, Cecilia Eydoux, Gilles Querat, Xavier de Lamballerie, Bruno Canard, Karine Alvarez, Jean-Claude Guillemot, Karine Barral
European Journal of Medicinal Chemistry 109:146-156 (2016)10.1016/j.ejmech.2015.12.046
Exploring Selective Inhibition of the First Bromodomain of the Human Bromodomain and Extra-terminal Domain (BET) Proteins
Brigitt Raux, Yuliia Voitovich, Carine Derviaux, Adrien Lugari, Etienne Rebuffet, Sabine Milhas, Stephane Priet, Thomas Roux, Eric Trinquet, Jean-Claude Guillemot, Stefan Knapp, Jean-Michel Brunel, Alexey Yu. Fedorov, Yves Collette, Philippe Roche, Stéphane Betzi, Sebastien Combes, Xavier Morelli
Journal of Medicinal Chemistry 59:1634-1641 (2016)10.1021/acs.jmedchem.5b01708
Structural characterization of the N-terminal part of the MERS-CoV nucleocapsid by X-ray diffraction and small-angle X-ray scattering
Nicolas Papageorgiou, Julie Lichiere, Amal Baklouti, Francois Ferron, Marion Sevajol, Bruno Canard, Bruno Coutard
Acta crystallographica Section D : Structural biology [1993-...] 72:192-202 (2016)10.1107/S2059798315024328
How disordered is my protein and what is its disorder for? A guide through the “dark side” of the protein universe
Philippe Lieutaud, François Ferron, Alexey V Uversky, Lukasz Kurgan, Vladimir N Uversky, Sonia Longhi
Intrinsically Disordered Proteins 4:e1259708 (2016)10.1080/21690707.2016.1259708
The viral capping enzyme nsP1: a novel target for the inhibition of chikungunya virus infection
L. Delang, C. Li, A. Tas, G. Querat, I. C. Albulescu, T. de Burghgraeve, N. A. Segura Guerrero, A. Gigante, G. Piorkowski, Etienne Decroly, D. Jochmans, B. Canard, E. J. Snijder, M. J. Perez-Perez, M. J. Van Hemert, B. Coutard, P. Leyssen, J. Neyts
Scientific Reports 6:31819 (2016)10.1038/srep31819
Involvement of an Arginine Triplet in M1 Matrix Protein Interaction with Membranes and in M1 Recruitment into Virus-Like Particles of the Influenza A(H1N1)pdm09 Virus
Adeline Kerviel, Shantoshini Dash, Olivier Moncorgé, Baptiste Panthu, Jan Prchal, Didier Decimo, Théophile Ohlmann, Bruno Lina, Cyril Favard, Etienne Decroly, Michèle Ottmann, Philippe Roingeard, Delphine Muriaux
PLoS ONE 11:e0165421 (2016)10.1371/journal.pone.0165421
Chemical diversity and antiviral potential in the pantropical Diospyros genus
Laure-Anne Peyrat, Véronique Eparvier, Cécilia Eydoux, Jean-Claude Guillemot, Didier Stien, Marc Litaudon
Fitoterapia (2016)10.1016/j.fitote.2016.04.017
Synthesis and structural characterization of monomeric and dimeric peptide nucleic acids prepared by using microwave-promoted multicomponent reactions
Reuben Ovadia, Aurelien Lebrun, Ivan Barvik, Jean-Jacques Vasseur, Carine Baraguey, Karine Alvarez
Organic & Biomolecular Chemistry 13:11052-11071 (2015)10.1039/c5ob01604e
Arenavirus Glycan Shield Promotes Neutralizing Antibody Evasion and Protracted Infection
Rami Sommerstein, Lukas Flatz, Melissa M. Remy, Pauline Malinge, Giovanni Magistrelli, Nicolas Fischer, Mehmet Sahin, Andreas Bergthaler, Sebastien Igonet, Jan Ter Meulen, Dorothee Rigo, Paolo Meda, Nadia Rabah, Bruno Coutard, Thomas A. Bowden, Paul-Henri Lambert, Claire-Anne Siegrist, Daniel D. Pinschewer
PLoS Pathogens 11 (2015)10.1371/journal.ppat.1005276
NMR study of non-structural proteins-part II: H-1, C-13, N-15 backbone and side-chain resonance assignment of macro domain from Venezuelan equine encephalitis virus (VEEV)
Garyfallia I. Makrynitsa, Dioni Ntonti, Konstantinos D. Marousis, Aikaterini C. Tsika, Julie Lichiere, Nicolas Papageorgiou, Bruno Coutard, Detlef Bentrop, Georgios A. Spyroulias
Biomolecular NMR Assignments 9:247-251 (2015)10.1007/s12104-014-9584-9
mRNA Capping by Venezuelan Equine Encephalitis Virus nsP1: Functional Characterization and Implications for Antiviral Research
Changqing Li, Jaime Guillen, Nadia Rabah, Alexandre Blanjoie, Francoise Debart, Jean-Jacques Vasseur, Bruno Canard, Etienne Decroly, Bruno Coutard
Journal of Virology 89:8292-8303 (2015)10.1128/JVI.00599-15
Enzymatic synthesis of acyclic nucleoside thiophosphonate diphosphates: Effect of the alpha-phosphorus configuration on HIV-1 RT activity
Stephane Priet, Loic Roux, Magali Saez-Ayala, Francois Ferron, Bruno Canard, Karine Alvarez
Antiviral Research 117:122-131 (2015)10.1016/j.antiviral.2015.03.003
NMR study of non-structural proteins-part I: H-1, C-13, N-15 backbone and side-chain resonance assignment of macro domain from Mayaro virus (MAYV)
Efstathios Melekis, Aikaterini C. Tsika, Julie Lichiere, Christos T. Chasapis, Irene Margiolaki, Nicolas Papageorgiou, Bruno Coutard, Detlef Bentrop, Georgios A. Spyroulias
Biomolecular NMR Assignments 9:191-195 (2015)10.1007/s12104-014-9572-0
Structural and biophysical analysis of sequence insertions in the Venezuelan Equine Encephalitis Virus macro domain
Jaime Guillen, Julie Lichiere, Nadia Rabah, Brett F. Beitzel, Bruno Canard, Bruno Coutard
Virus Research 201:94-100 (2015)10.1016/j.virusres.2015.02.018
Primer-Dependent and Primer-Independent Initiation of Double Stranded RNA Synthesis by Purified Arabidopsis RNA-Dependent RNA Polymerases RDR2 and RDR6
Anthony Devert, Nicolas Fabre, Maïna Floris, Bruno Canard, Christophe Robaglia, Patrice Crété
PLoS ONE 10 (2015)10.1371/journal.pone.0120100.s006
Les années 2010
Michel Bouvier, Herve Chneiweiss, Bertrand Jordan, Gilbert Lagrue, Joël Ménard, Jean-Luc Teillaud, Guido Kroemer, Anne-Marie Moulin, Antoine Flahault, Laurence Lafanechère, Eric Maréchal, Jean-Claude Guillemot, Marie Gaille
Médecine/Sciences 31:91-111 (2015)10.1051/medsci/201531s1006
The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family
Lonneke van Der Linden, Laia Vives-Adrián, Barbara Selisko, Cristina Ferrer-Orta, Xinran Liu, Kjerstin Lanke, Rachel Ulferts, Armando M. de Palma, Federica Tanchis, Nesya Goris, David Lefebvre, Kris de Clercq, Pieter Leyssen, Céline Lacroix, Gerhard Pürstinger, Bruno Coutard, Bruno Canard, David D. Boehr, Jamie J. Arnold, Craig E. Cameron, Nuria Verdaguer, Johan Neyts, Frank J. M van Kuppeveld
PLoS Pathogens 11 (2015)10.1371/journal.ppat.1004733.s010
X-ray structure and activities of an essential Mononegavirales L-protein domain
Guido C. Paesen, Axelle Collet, Corinne Sallamand, Francoise Debart, Jean-Jacques Vasseur, Bruno Canard, Etienne Decroly, Jonathan M. Grimes
Nature Communications 6:8749 (2015)10.1038/ncomms9749
Modulation of violacein production and phenotypes associated with biofilm by exogenous quorum sensing N-acylhomoserine lactones in the marine bacterium Pseudoalteromonas ulvae TC14
Armande Mireille Ayé, Maryse Bonnin-Jusserand, Florence Brian-Jaisson, Annick Ortalo-Magné, Gérald Culioli, Rose Koffi Nevry, Nadia Rabah, Yves Blache, Maëlle Molmeret
Microbiology 161:2039-2052 (2015)10.1099/mic.0.000147
La chémobiologie : stratégies transfrontalières
Laurence Lafanechère, Eric Maréchal, Jean-Claude Guillemot
Médecine/Sciences 30:2 (2014)10.1051/medsci/20143012001
Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus
Marion Sevajol, Lorenzo Subissi, Etienne Decroly, Bruno Canard, Isabelle Imbert
Virus Research 194:90-99 (2014)10.1016/j.virusres.2014.10.008
The methyltransferase domain of dengue virus protein NS5 ensures efficient RNA synthesis initiation and elongation by the polymerase domain
Supanee Potisopon, Stéphane Priet, Axelle Collet, Etienne Decroly, Bruno Canard, Barbara Selisko
Nucleic Acids Research 42 (2014)10.1093/nar/gku666
One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities
Lorenzo Subissi, Clara Posthuma, Axelle Collet, Jessika Zevenhoven-Dobbe, Alexander Gorbalenya, Etienne Decroly, Eric Snijder, Bruno Canard, Isabelle Imbert
Proceedings of the National Academy of Sciences of the United States of America 111 (2014)10.1073/pnas.1323705111
Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes
Mickaël Bouvet, Adrien Lugari, Clara C. Posthuma, Jessika C. Zevenhoven, Stéphanie Bernard, Stéphane Betzi, Isabelle Imbert, Bruno Canard, Jean-Claude Guillemot, Patrick Lécine, Susanne Pfefferle, Christian Drosten, Eric J. Snijder, Etienne Decroly, Xavier Morelli
Journal of Biological Chemistry 289:25783-25796 (2014)10.1074/jbc.M114.577353
Identification of a Src kinase SH3 binding site in the C-terminal domain of the human ErbB2 receptor tyrosine kinase.
Olivier Bornet, Matthieu Nouailler, Michaël Feracci, Corinne Sebban-Kreuzer, Deborah Byrne, Hubert Halimi, Xavier Morelli, Ali Badache, Francoise Guerlesquin
FEBS Letters 588:2031-6 (2014)10.1016/j.febslet.2014.04.029
Assessment of Dengue virus helicase and methyltransferase as targets for fragment-based drug discovery
Bruno Coutard, Etienne Decroly, Changqing Li, Andrew Sharff, Julien Lescar, Gérard Bricogne, Karine Barral
Antiviral Research 106:61-70 (2014)10.1016/j.antiviral.2014.03.013
Biodiversity as a source of potent and selective inhibitors of Chikungunya virus replication
Pieter Leyssen, Jacqueline Smadja, Philippe Rasoanaivo, Ameenah Gurib-Fakim, Mohamad Fawzi-Mahomoodally, Bruno Canard, Jean-Claude Guillemot, Marc Litaudon, Françoise Guéritte
151-160 (2014)10.1002/9781118460566.ch11
Atomic resolution description of the interaction between the nucleoprotein and phosphoprotein of Hendra virus.
Guillaume Communie, Johnny Habchi, Filip Yabukarski, David Blocquel, Robert Schneider, Nicolas Tarbouriech, Nicolas Papageorgiou, Rob W H Ruigrok, Marc Jamin, Malene Ringkjøbing Jensen, Sonia Longhi, Martin Blackledge
PLoS Pathogens 9:e1003631 (2013)
Development of specific dengue virus 2'-O- and N7-methyltransferase assays for antiviral drug screening.
Karine Barral, Corinne Sallamand, Christiane Petzold, Bruno Coutard, Axelle Collet, Yann Thillier, Jan Zimmermann, Jean-Jacques Vasseur, Bruno Canard, Jacques Rohayem, Françoise Debart, Etienne Decroly
Antiviral Research 99:292-300 (2013)10.1016/j.antiviral.2013.06.001
Plasma Peptide Biomarker Discovery for Amyotrophic Lateral Sclerosis by MALDI –TOF Mass Spectrometry Profiling
Laurence Conraux, Catherine Pech, Halim Guerraoui, Denis Loyaux, Pascual Ferrara, Jean-Claude Guillemot, Vincent Meininger, Pierre-François Pradat, François Salachas, Gaelle Bruneteau, Nadine Le Forestier, Lucette Lacomblez
PLoS ONE 8:e79733 (2013)10.1371/journal.pone.0079733
Structural Basis for Galectin-1-dependent Pre-B Cell Receptor (Pre-BCR) Activation
Latifa Elantak, Marion Espeli, Annie Boned, Olivier Bornet, Jeremy Bonzi, Laurent Gauthier, Mikael Feracci, Philippe Roche, Francoise Guerlesquin, Claudine Schiff
Journal of Biological Chemistry 287:44703-44713 (2012)10.1074/jbc.m112.395152
Antiviral chlorinated daphnane diterpenoid orthoesters from the bark and wood of Trigonostemon cherrieri.
Pierre-Marie Allard, Pieter Leyssen, Marie-Thérèse Martin, Mélanie Bourjot, Vincent Dumontet, Cécilia Eydoux, Jean-Claude Guillemot, Bruno Canard, Cyril Poullain, Françoise Guéritte, Marc Litaudon
Phytochemistry 84:160-8 (2012)10.1016/j.phytochem.2012.07.023
Structural basis for galectin-1-dependent pre-B cell receptor (pre-BCR) activation.
Latifa El Antak, Marion Espeli, Annie Boned, Olivier Bornet, Jeremy Bonzi, Laurent Gauthier, Mikael Feracci, Philippe Roche, Françoise Guerlesquin, Claudine Schiff
Journal of Biological Chemistry 287:44703-13 (2012)10.1074/jbc.M112.395152
Chemical constituents of Anacolosa pervilleana and their antiviral activities.
Mélanie Bourjot, Pieter Leyssen, Cécilia Eydoux, Jean-Claude Guillemot, Bruno Canard, Philippe Rasoanaivo, Françoise Guéritte, Marc Litaudon
Fitoterapia 83:1076-1080 (2012)10.1016/j.fitote.2012.05.004
Flacourtosides A-F, Phenolic Glycosides Isolated from Flacourtia ramontchi.
Mélanie Bourjot, Pieter Leyssen, Cécilia Eydoux, Jean-Claude Guillemot, Bruno Canard, Philippe Rasoanaivo, Françoise Guéritte, Marc Litaudon
Journal of Natural Products 75:752-8 (2012)10.1021/np300059n
SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates
H. Lahouassa, W. Daddacha, H. Hofmann, D. Ayinde, E. C. Logue, L. Dragin, N. Bloch, C. Maudet, M. Bertrand, T. Gramberg, G. Pancino, S. Priet, B. Canard, Nadine Laguette, M. Benkirane, C. Transy, Nr Landau, B. Kim, F. Margottin-Goguet
Nature Immunology 13:223-8. doi: 10.1038/ni.2236 (2012)10.1038/ni.2236
Validation of an Image Analysis Method for the Determination of the Parasitemia in Malaria-Infected Erythrocyte Cultures
German Gonzalez, Karine Alvarez, Mirko Zimic, Stephane Bertani, Michel Sauvain, Eric Deharo
(2012)
Synthesis of 5' cap-0 and cap-1 RNAs using solid-phase chemistry coupled with enzymatic methylation by human (guanine-N 7)-methyl transferase
Yann Thillier, Etienne Decroly, François Morvan, Bruno Canard, Jean-Jacques Vasseur, Françoise Debart
RNA 18:856-868 (2012)10.1261/rna.030932.111
Molecular Basis for Nucleotide Conservation at the Ends of the Dengue Virus Genome
Barbara Selisko, Supanee Potisopon, Rym Agred, Stephane Priet, Isabelle Varlet, Yann Thillier, Corinne Sallamand, Françoise Debart, Jean-Jacques Vasseur, Bruno Canard
PLoS Pathogens 8:e1002912 (2012)10.1371/journal.ppat.1002912
Single pH buffer refolding screen for protein from inclusion bodies
Bruno Coutard, Etienne Danchin, Rachid Oubelaid, Bruno Canard, Christophe Bignon
Protein Expression and Purification 82:352-359 (2012)10.1016/j.pep.2012.01.014
Alkylated flavanones from the bark of Cryptocarya chartacea as dengue virus NS5 polymerase inhibitors.
Pierre-Marie Allard, Elise Tran Huu Dau, Cécilia Eydoux, Jean-Claude Guillemot, Vincent Dumontet, Cyril Poullain, Bruno Canard, Françoise Guéritte, Marc Litaudon
Journal of Natural Products 74:2446-53 (2011)10.1021/np200715v
MEMO associated with an ErbB2 receptor phosphopeptide reveals a new phosphotyrosine motif.
Mikael Feracci, Cyril Pimentel, Olivier Bornet, Philippe Roche, Danièle Salaün, Ali Badache, Françoise Guerlesquin
FEBS Letters 585:2688-92 (2011)10.1016/j.febslet.2011.07.048
Acyclic nucleoside thiophosphonates as potent inhibitors of HIV and HBV replication
Karine Barral, Clément Weck, Nadine Payrot, Loic Roux, Céline Durafour, Fabien Zoulim, Johan Neyts, Jan Balzarini, Bruno Canard, Stephane Priet, Karine Alvarez
European Journal of Medicinal Chemistry 46:4281-4288 (2011)10.1016/j.ejmech.2011.06.034
Crystallization and diffraction analysis of the SARS coronavirus nsp10-nsp16 complex.
Claire Debarnot, Isabelle Imbert, Francois Ferron, Laure Gluais, Isabelle Varlet, Nicolas Papageorgiou, Mickaël Bouvet, Julien Lescar, Etienne Decroly, Bruno Canard
Acta crystallographica Section F : Structural biology communications [2014-...] 67:404-8 (2011)10.1107/S1744309111002867
PHYTOCHIK: Biodiversity As A Source of Selective Inhibitors of CHIKV Replication
Pieter Leyssen, Marc Litaudon, Jean-Claude Guillemot, Philippe Rasoanaivo, Jacqueline Smadja, Ameenah Gurib-Fakim, Bruno Canard, Françoise Guéritte
Antiviral Research 90:A36-A37 (2011)10.1016/j.antiviral.2011.03.048
Flavivirus NS3 and NS5 proteins interaction network: a high-throughput yeast two-hybrid screen.
Marc Le Breton, Laurène Meyniel-Schicklin, Alexandre Deloire, Bruno Coutard, Bruno Canard, Xavier de Lamballerie, Patrice Andre, Chantal Rabourdin-Combe, Vincent Lotteau, Nathalie Davoust
BMC Microbiology 11:234 (2011)10.1186/1471-2180-11-234
Genomics and structure/function studies of Rhabdoviridae proteins involved in replication and transcription.
R. Assenberg, O Delmas, B Morin, C Graham, X de Lamballerie, C Laubert, B Coutard, J Grimes, J Neyts, R J Owens, B Brandt, A Gorbalenya, P Tucker, D I Stuart, Bruno Canard, Hervé Bourhy
Antiviral Research 87:149-61 (2010)10.1016/j.antiviral.2010.02.322
Synthesis and antiviral activity of boranophosphonate isosteres of AZT and d4T monophosphates
Karine Barral, Stephane Priet, Céline de Michelis, Joséphine Sire, Johan Neyts, Jan Balzarini, Bruno Canard, Karine Alvarez
European Journal of Medicinal Chemistry 45:849-856 (2010)10.1016/j.ejmech.2009.11.012
The 2C putative helicase of echovirus 30 adopts a hexameric ring-shaped structure
N. Papageorgiou, B. Coutard, V. Lantez, E. Gautron, Olivier Chauvet, C. Baronti, H. Norder, X. de Lamballerie, V. Heresanu, N. Ferte, S. Veesler, A.E. Gorbalenya, B. Canard
Acta crystallographica Section D : Structural biology [1993-...] 66:1116-1120 (2010)
3'-Deoxy phosphoramidate dinucleosides as improved inhibitors of Hepatitis C virus subgenomic replicon and NS5B polymerase activity.
Stephane Priet, Ivan Zlatev, Ivan Barvik, Katrien Geerts, Pieter Leyssen, Johan Neyts, Hélène Dutartre, Bruno Canard, Jean-Jacques Vasseur, François Morvan, Karine Alvarez
Journal of Medicinal Chemistry 53:6608-6617 (2010)10.1021/jm100102v
NMR and MD investigations of human galectin-1/oligosaccharide complexes.
Christophe Meynier, Mikael Feracci, Marion Espeli, Florence Chaspoul, Philippe Gallice, Claudine Schiff, Françoise Guerlesquin, Roche Philippe
Biophysical Journal 97:3168-77 (2009)10.1016/j.bpj.2009.09.026
Identification of allosteric inhibitors blocking the hepatitis C virus polymerase NS5B in the RNA synthesis initiation step.
Stéphane Betzi, Cécilia Eydoux, Cécile Bussetta, Marilyne Blemont, Pieter Leyssen, Claire Debarnot, Mohamed Ben-Rahou, Jacques Haiech, Marcel Hibert, Françoise Gueritte, David S. Grierson, Jean-Louis Romette, Jean-Claude Guillemot, Johan Neyts, Karine Alvarez, Xavier Morelli, Hélène Dutartre, Bruno Canard
Antiviral Research 84:48-59 (2009)10.1016/j.antiviral.2009.06.009
The Crystal Structures of Chikungunya and Venezuelan Equine Encephalitis Virus nsP3 Macro Domains Define a Conserved Adenosine Binding Pocket
H. Malet, B. Coutard, S. Jamal, H. Dutartre, N. Papageorgiou, M. Neuvonen, T. Ahola, N. Forrester, E. A. Gould, D. Lafitte, F. Ferron, J. Lescar, A. Gorbalenya, X. de Lamballerie, B. Canard
Journal of Virology 83:6534-6545 (2009)10.1128/JVI.00189-09
A pyrazolotriazolopyrimidinamine inhibitor of bovine viral diarrhea virus replication that targets the viral RNA-dependent RNA polymerase
Jan Paeshuyse, Carine Letellier, Matheus Froeyen, Hélène Dutartre, Robert Vrancken, Bruno Canard, Erik de Clercq, Alain Gueiffier, Jean-Claude Teulade, Piet Herdewijn, Gerhard Puerstinger, Frank Koenen, Pierre Kerkhofs, Pier Giovanni Baraldi, Johan Neyts
Antiviral Research 82:141-147 (2009)10.1016/j.antiviral.2009.02.192
Structure of Human Pancreatic Lipase-Related Protein 2 with the Lid in an Open Conformation ,
Cécilia Eydoux, Silvia Spinelli, Tara Davis, John Walker, Alma Seitova, Sirano Dhe-Paganon, Alain de Caro, Christian Cambillau, Frédéric Carrière
Biochemistry 47:9553-9564 (2008)10.1021/bi8005576
Occurrence of pancreatic lipase-related protein-2 in various species and its relationship with herbivore diet
Josiane de Caro, Cécilia Eydoux, Slim Chérif, Régine Lebrun, Youssef Gargouri, Frédéric Carrière, Alain de Caro
Comparative Biochemistry and Physiology. Part B, Biochemistry and Molecular Biology 150:1-9 (2008)10.1016/j.cbpb.2008.01.007
Acyclic phosphonate nucleotides and human adenylate kinases: impact of a borano group on alpha-P position.
D. Topalis, K. Alvarez, K. Barral, Hélène Munier-Lehmann, B. Schneider, M. Véron, C. Guerreiro, Laurence A Mulard, C. El-Amri, B. Canard, Dominique Deville-Bonne
Nucleosides, Nucleotides and Nucleic Acids 27:319-31 (2008)10.1080/15257770801941952
Carboxyl-Terminal Disulfide Bond of Acid Sphingomyelinase Is Critical for Its Secretion and Enzymatic Function
Ching Yin Lee, Taku Tamura, Nadia Rabah, Dong-Young Donna Lee, Isabelle Ruel, Anouar Hafiane, Iulia Iatan, Dana Nyholt, Frédéric Laporte, Claude Lazure, Ikuo Wada, Larbi Krimbou, Jacques Genest
Biochemistry 46:14969-14978 (2007)10.1021/bi700817g
The Imidazopyrrolopyridine Analogue AG110 Is a Novel, Highly Selective Inhibitor of Pestiviruses That Targets the Viral RNA-Dependent RNA Polymerase at a Hot Spot for Inhibition of Viral Replication
Jan Paeshuyse, Jean-Michel Chezal, Matheus Froeyen, Pieter Leyssen, Hélène Dutartre, Robert Vrancken, Bruno Canard, Carine Letellier, Tong Li, Harald Mittendorfer, Frank Koenen, Pierre Kerkhofs, Erik de Clercq, Piet Herdewijn, Gerhard Puerstinger, Alain Gueiffier, Olivier Chavignon, Jean-Claude Teulade, Johan Neyts
Journal of Virology 81:11046-11053 (2007)10.1128/JVI.00388-07
In Vitro Suppression of K65R Reverse Transcriptase-Mediated Tenofovir- and Adefovir-5'-Diphosphate Resistance Conferred by the Boranophosphonate Derivatives
A. Frangeul, Karine Barral, K. Alvarez, B. Canard
Antimicrobial Agents and Chemotherapy 51:3162-3167 (2007)10.1128/AAC.00145-07
Further biochemical characterization of human pancreatic lipase-related protein 2 expressed in yeast cells
Cécilia Eydoux, Josiane de Caro, Francine Ferrato, Paul Boullanger, Dominique Lafont, René Laugier, Frédéric Carrière, Alain de Caro
Journal of Lipid Research 48:1539-1549 (2007)10.1194/jlr.M600486-JLR200
The C-terminal region of the proprotein convertase 1/3 (PC1/3) exerts a bimodal regulation of the enzyme activity in vitro
Nadia Rabah, Dany Gauthier, Jimmy Dikeakos, Timothy Reudelhuber, Claude Lazure
FEBS Journal 274:3482-3491 (2007)10.1111/j.1742-4658.2007.05883.x
Crystal Structure of the RNA Polymerase Domain of the West Nile Virus Non-structural Protein 5
Hélène Malet, Marie-Pierre Egloff, Barbara Selisko, Rebecca E Butcher, Peter J Wright, Michael Roberts, Arnaud Gruez, Gerlind Sulzenbacher, Clemens Vonrhein, Gérard Bricogne, Jason M Mackenzie, Alexander A Khromykh, Andrew D Davidson, Bruno Canard
Journal of Biological Chemistry (2007)10.1074/jbc.M607273200
High-yield production of short GpppA- and 7MeGpppA-capped RNAs and HPLC-monitoring of methyltransfer reactions at the guanine-N7 and adenosine-2'-O positions.
F. Peyrane, B. Selisko, Etienne Decroly, Jean-Jacques Vasseur, D. Benarroch, B. Canard, Karine Alvarez
Nucleic Acids Research 35:1-11 (2007)10.1093/nar/gkl1119
Preparation of purine and pyrimidine acyclic nucleotide analogs as antiviral agents
Karine Barral, Bruno Canard, Karine Alvarez, Jean-Louis Romette, Johan Neyts, Jan Balzarini
(2007)
High-yield production of short GppA- and 7Me GppA-capped RNAs and HPLC-monitoring of methyltransfer reactions at the guanine-N7 and adenosine-2'O positions.
F. Peyrane, B. Selisko, Etienne Decroly, J.-J. Vasseur, D. Benarroch, B. Canard, K. Alvarez
Nucleic Acids Research 35:1-11 (2007)
Synthesis, in Vitro Antiviral Evaluation, and Stability Studies of Novel α-Borano-Nucleotide Analogues of 9-[2-(Phosphonomethoxy)ethyl]adenine and ( R )-9-[2-(Phosphonomethoxy)propyl]adenine
Karine Barral, Stephane Priet, Joséphine Sire, Johan Neyts, Jan Balzarini, Bruno Canard, Karine Alvarez
Journal of Medicinal Chemistry 49:7799-7806 (2006)10.1021/jm060030y
Interaction of human immunodeficiency virus type 1 Vif with Gag and Gag-Pol precursors: co-encapsidation and interference with viral protease-mediated Gag processing.
Marion Bardy, B. Gay, S. Pebernard, N. Chazal, M. Courcoul, R. Vigne, Etienne Decroly, P. Boulanger
(2006)
General Catalytic Deficiency of Hepatitis C Virus RNA Polymerase with an S282T Mutation and Mutually Exclusive Resistance towards 2'-Modified Nucleotide Analogues
Hélène Dutartre, Cécile Bussetta, Joëlle Boretto, Bruno Canard
Antimicrobial Agents and Chemotherapy 50:4161-4169 (2006)10.1128/AAC.00433-06
Human pancreatic lipase-related protein 2: Tissular localization along the digestive tract and quantification in pancreatic juice using a specific ELISA
Cécilia Eydoux, Ahmed Aloulou, Josiane de Caro, Philippe Grandval, René Laugier, Frédéric Carrière, Alain de Caro
Biochimica et Biophysica Acta (BBA) - General Subjects 1760:1497-1504 (2006)10.1016/j.bbagen.2006.06.005
SPINE bioinformatics and data-management aspects of high-throughput structural biology.
S. Albeck, Pedro Maria Alzari, C. Andreini, L. Banci, I. M. Berry, I. Bertini, C. Cambillau, B. Canard, L. Carter, S. X. Cohen, J. M. Diprose, O. Dym, R. M. Esnouf, C. Felder, F. Ferron, F. Guillemot, R. Hamer, M. Ben Jelloul, R. A. Laskowski, T. Laurent, S. Longhi, R. Lopez, C. Luchinat, H. Malet, T. Mochel, R. J. Morris, L. Moulinier, T. Oinn, A. Pajon, Y. Peleg, A. Perrakis, O. Poch, J. Prilusky, A. Rachedi, R. Ripp, A. Rosato, I. Silman, D. I. Stuart, J. L. Sussman, J. C. Thierry, J. D. Thompson, J. M. Thornton, T. Unger, B. Vaughan, W. Vranken, J. D. Watson, G. Whamond, K. Henrick
Acta crystallographica Section D : Structural biology [1993-...] 62:1184-95 (2006)10.1107/S090744490602991X
Application of the use of high-throughput technologies to the determination of protein structures of bacterial and viral pathogens
M. J. Fogg, P. Alzari, M. Bahar, I. Bertini, J.-M. Betton, W. P. Burmeister, C. Cambillau, B. Canard, M. Carrondo, M. Coll, S. Daenke, O. Dym, M.-P. Egloff, F. J. Enguita, A. Geerlof, A. Haouz, T. A. Jones, Q. Ma, S. N. Manicka, M. Migliardi, P. Nordlund, R. J. Owens, Y. Peleg, G. Schneider, R. Schnell, D. I. Stuart, N. Tarbouriech, T. Unge, A. J. Wilkinson, M. Wilmanns, K. S. Wilson, O. Zimhony, J. M. Grimes
Acta crystallographica Section D : Structural biology [1993-...] 62:1196-1207 (2006)10.1107/S0907444906030915
Structural and Functional Basis for ADP-Ribose and Poly(ADP-Ribose) Binding by Viral Macro Domains
Marie-Pierre Egloff, Hélène Malet, Ákos Putics, Maarit Heinonen, Hélène Dutartre, Antoine Frangeul, Arnaud Gruez, Valérie Campanacci, Christian Cambillau, John Ziebuhr, Tero Ahola, Bruno Canard
Journal of Virology 80:8493-8502 (2006)10.1128/JVI.00713-06
Comparative mechanistic studies of de novo RNA synthesis by flavivirus RNA-dependent RNA polymerases
Barbara Selisko, Hélène Dutartre, Jean-Claude Guillemot, Claire Debarnot, Delphine Benarroch, Alexander Khromykh, Philippe Desprès, Marie-Pierre Egloff, Bruno Canard
Virology 351:145-158 (2006)10.1016/j.virol.2006.03.026
Expression, purification and crystallization of the SARS-CoV macro domain
Hélène Malet, Karen Dalle, Nicolas Bremond, Fabienne Tocque, Stéphanie Blangy, Valérie Campanacci, Bruno Coutard, Sacha Grisel, Julie Lichiere, Violaine Lantez, Christian Cambillau, Bruno Canard, Marie-Pierre Egloff
Acta Crystallographica Section F: Structural Biology and Crystallization Communications 62:405-408 (2006)10.1107/S1744309106009274
Synthesis and Anti-BVDV Activity of Acridones As New Potential Antiviral Agents 1
Oriana Tabarrini, Giuseppe Manfroni, Arnaldo Fravolini, Violetta Cecchetti, Stefano Sabatini, Erik de Clercq, Jef Rozenski, Bruno Canard, Hélène Dutartre, Jan Paeshuyse, Johan Neyts
Journal of Medicinal Chemistry 49:2621-2627 (2006)10.1021/jm051250z
Single Amino Acid Substitution in the PC1/3 Propeptide Can Induce Significant Modifications of Its Inhibitory Profile toward Its Cognate Enzyme
Nadia Rabah, Dany Gauthier, Brian Wilkes, Daniel Gauthier, Claude Lazure
Journal of Biological Chemistry 281:7556-7567 (2006)10.1074/jbc.M510607200
A Novel, Highly Selective Inhibitor of Pestivirus Replication That Targets the Viral RNA-Dependent RNA Polymerase
Jan Paeshuyse, Pieter Leyssen, Eric Mabery, Nina Boddeker, Robert Vrancken, Matheus Froeyen, Israrul Ansari, Hélène Dutartre, Jef Rozenski, Laura Gil, Carine Letellier, Robert Lanford, Bruno Canard, Frank Koenen, Pierre Kerkhofs, Ruben Donis, Piet Herdewijn, Julia Watson, Erik de Clercq, Gerhard Puerstinger, Johan Neyts
Journal of Virology 80:149-160 (2005)10.1128/JVI.80.1.149-160.2006
Borano-nucleotides: new analogues to circumvent HIV-1 RT-mediated nucleoside drug-resistance
Karine Alvarez, Jérôme Deval, Boulbaba Selmi, Karine Barral, Joelle Boretto, Catherine Guerreiro, Laurence S Mulard, Robert Sarfati, Bruno Canard
Nucleosides, Nucleotides and Nucleic Acids 24:419-422 (2005)10.1081/NCN-200059951
VaZyMolO: a tool to define and classify modularity in viral proteins
François Ferron, Corinne Rancurel, Sonia Longhi, Christian Cambillau, Bernard Henrissat, Bruno Canard
Journal of General Virology 86:743-749 (2005)10.1099/vir.0.80590-0
A Relaxed Discrimination of 2′- O -Methyl-GTP Relative to GTP between de Novo and Elongative RNA Synthesis by the Hepatitis C RNA-dependent RNA Polymerase NS5B
Hélène Dutartre, Joëlle Boretto, Jean Claude Guillemot, Bruno Canard
Journal of Biological Chemistry 280:6359-6368 (2005)10.1074/jbc.M410191200
The modeled structure of the RNA dependent RNA polymerase of GBV-C virus suggests a role for motif E in Flaviviridae RNA polymerases.
Francois Ferron, Cécile Bussetta, Hélène Dutartre, Bruno Canard
BMC Bioinformatics 6:255 (2005)10.1186/1471-2105-6-255
Utilization of a new biotinylation reagent in the development of a nondiscriminatory investigative approach for the study of cell surface proteins
Daniel Gauthier, Bernard Gibbs, Nadia Rabah, Claude Lazure
Proteomics 4:3783-3790 (2004)10.1002/pmic.200300860
Improved PC1/3 production through recombinant expression in insect cells and larvae
Nadia Rabah, Daniel Gauthier, Dany Gauthier, Claude Lazure
Protein Expression and Purification 37:377-384 (2004)10.1016/j.pep.2004.06.014
The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world
Marie-Pierre Egloff, Francois Ferron, Valérie Campanacci, Sonia Longhi, Corinne Rancurel, Hélène Dutartre, Eric J Snijder, Alexander Gorbalenya, Christian Cambillau, Bruno Canard
Proceedings of the National Academy of Sciences of the United States of America 101:3792-3796 (2004)10.1073/pnas.0307877101
Crystal Structure of E. coli Alcohol Dehydrogenase YqhD: Evidence of a Covalently Modified NADP Coenzyme
Gerlind Sulzenbacher, Karine Alvarez, Robert H H van den Heuvel, Cees Versluis, Silvia Spinelli, Valé Rie Campanacci, Christel Valencia, Christian Cambillau, Hans Eklund, Mariella Tegoni
Journal of Molecular Biology 342:489-502 (2004)10.1016/j.jmb.2004.07.034
Structural genomics of the SARS coronavirus: cloning, expression, crystallization and preliminary crystallographic study of the Nsp9 protein
Valérie Campanacci, Marie-Pierre Egloff, Sonia Longhi, François Ferron, Corinne Rancurel, Aurelia Salomoni, Cécile Durousseau, Fabienne Tocque, Nicolas Brémond, Jessika Dobbe, Eric Snijder, Bruno Canard, Christian Cambillau
Acta Crystallographica Section D: Biological Crystallography 59:1628-1631 (2003)10.1107/S0907444903016779
Barley Serine Proteinase Inhibitor 2-Derived Cyclic Peptides as Potent and Selective Inhibitors of Convertases PC1/3 and Furin
Michèle Villemure, Alain Fournier, Dany Gauthier, Nadia Rabah, Brian Wilkes, Claude Lazure
Biochemistry 42:9659-9668 (2003)10.1021/bi034418w
Structural Analysis of the Activation of Ribavirin Analogs by NDP Kinase: Comparison with Other Ribavirin Targets
Sarah Gallois-Montbrun, Yuxing Chen, Hélène Dutartre, Magali Sophys, Solange Moréra, Catherine Guerreiro, Benoit Schneider, Laurence S Mulard, Joël Janin, Michel Véron, Dominique Deville-Bonne, Bruno Canard
Molecular Pharmacology 63:538-546 (2003)10.1124/mol.63.3.538
Porcine odorant-binding protein selectively binds to a human olfactory receptor
Valéry Matarazzo, Nicole Zsürger, Jean-Claude Guillemot, Olivier Clot-Faybesse, Jean-Marie Botto, Claude Dal Farra, Marc Crowe, Jacques Demaille, Jean-Pierre Vincent, Jean Mazella, Catherine Ronin
Chemical Senses 27:691-701 (2002)10.1093/chemse/27.8.691
Identification of two tamoxifen target proteins by photolabeling with 4-(2-morpholinoethoxy)benzophenone.
Fabienne Mésange, Mohamed Sebbar, Joël Capdevielle, Jean-Claude Guillemot, Pascual Ferrara, Francis Bayard, Marc E. Poirot, Jean-Charles Faye
Bioconjugate Chemistry 13:766-772 (2002)10.1021/bc015588t
Interaction of human immunodeficiency virus type 1 Vif with Gag and Gag-Pol precursors : co-encapsidation and interference with viral protease-mediated Gag processing
Martine Bardy, Bernard Gay, Stéphanie Pébernard, Nathalie Chazal, Marianne Courcoul, Robert Vigne, Etienne Decroly, Pierre Boulanger
Journal of General Virology 82:2719-2733 (2001)10.1099/0022-1317-82-11-2719
An Anti-Human Immunodeficiency Virus Multiple Antigen Peptide Encompassing the Cleavage Region of the Env Precursor Interferes With Membrane Fusion at a Post-CD4 Binding Step
Rym Barbouche, Etienne Decroly, Marie Paule Kieny, Emmanuel Fenouillet
Virology 273:169-177 (2000)10.1006/viro.2000.0368
Tyrosine phosphorylation of the Wiskott-Aldrich Syndrome protein by Lyn and Btk is regulated by CDC42
Rodolphe r. Guinamard, Pontus Aspenström, Michel Fougereau, Philippe Chavrier, Jean-Claude Guillemot
FEBS Letters 434:431-436 (1998)10.1016/s0014-5793(98)01016-3
The pro-oligonucleotide approach: solid phase synthesis and preliminary evaluation of model pro-dodecathymidylates
J.J. Vasseur, Guilhem Tosquellas, Karine Alvarez, Christelle Dell'Aquila, François Morvan, Jean-Jacques Vasseur, Jean-Louis Imbach, Bernard Rayner
Nucleic Acids Research 26:2069-2074 (1998)10.1093/nar/26.9.2069
Microsomal epoxide hydrolase of rat liver is a subunit of the anti-oestrogen-binding site.
Fabienne Mésange, Mohamed Sebbar, Blandine Kedjouar, Joël Capdevielle, Jean-Claude Guillemot, Pascual Ferrara, Francis Bayard, Frédéric Delarue, Jean-Charles Faye, Marc E. Poirot
Biochemical Journal 334 ( Pt 1):107-112 (1998)10.1042/bj3340107
Clostridium perfringens CPN50
Stewart Cole, Sei-Ichi Katayama, Bruno Dupuy, Thierry Garnier, Brigitte Saint-Joanis, Bruno Canard
645-647 (1998)10.1007/978-1-4615-6369-3_58
Mismatch repair in Xenopus egg extracts: DNA strand breaks act as signals rather than excision points
I. Varlet, B. Canard, P. Brooks, G. Cerovic, M. Radman
Proceedings of the National Academy of Sciences of the United States of America 93:10156-10161 (1996)10.1073/pnas.93.19.10156
Studies of recombinant amylosucrase
M. Remaud-Simeon, F. Albaret, B. Canard, I. Varlet, P. Colonna, R. M. Willemot, P. Monsan
313-320 (1995)
Rat platelets contain glycosylated and non-glycosylated forms of platelet factor 4. Identification and characterization by mass spectrometry.
Catherine Ravanat, Christian Gachet, Jean-Marc Herbert, Simone Schuhler, Jean-Claude Guillemot, Françoise Uzabiaga, Claudine Picard, Pascual Ferrara, Monique Freund, Jean-Pierre Cazenave
European Journal of Biochemistry 223:203-210 (1994)10.1111/j.1432-1033.1994.tb18984.x
Phylogenetic analysis of the pathogenic anaerobe Clostridium perfringens using the 16S rRNA nucleotide sequence.
Bruno Canard, Thierry Garnier, Bénédicte Lafay, Richard Christen, Stewart T. Cole
International Journal of Systematic Bacteriology 42:312-4 (1992)10.1099/00207713-42-2-312
Autolysis of Clostridium acetobutylicum ATCC 824
Christian Croux, B. Canard, G. Goma, Philippe Soucaille
Journal of general microbiology 138:861-869 (1992)
Purification and characterization of an extracellular muramidase of Clostridium acetobutylicum ATCC 824 that acts on non-n-acetylated pepticoglycan
Christian Croux, B. Canard, G. Goma, Philippe Soucaille
Applied and Environmental Microbiology 58:1075-1081 (1992)
Réseau
National
Européen
International
Financements
L’équipe tient à remercier ses soutiens institutionnels et privés










































